EXPERIMENTS: UV RADIATION EFFECTS on MOLECULES and CELLS
Kendric C. Smith
Emeritus Professor, Radiation Oncology (Radiation Biology)
Stanford University School of Medicine
800 Blossom Hill Road, Unit R169, Los Gatos, CA 95032
kendric@stanford.edu
web.stanford.edu/~kendric/
[Revised from his review: Ultraviolet Radiation Effects on Molecules and Cells, in The Science of Photobiology (1st Ed, 1977), (KC Smith, ed), Plenum Press, New York, pp.113-142.]
PHOTOCHEMISTRY OF NUCLEIC ACIDS
Hydrate Formation:
Determine the absorption spectrum (200-300 nm) of uridylic acid (30 µg/ml). Irradiate at 254 nm (15-W GE germicidal lamp) in a quartz or a UV-transparent plastic ("UVette") cuvette, and follow the disappearance of absorption with time at 260 nm. When the absorbance at 260 nm is less than about 0.2, determine the full absorption spectrum to see the chemical change in the uridylic acid (hydrate formation). Adjust the irradiated solution to ~pH 1 with concentrated HCl (~0.1 ml), cover and let stand at room temperature for 24 h, and determine the absorption spectrum to see the return of the absorption spectrum of uridylic acid (removal of the hydrate) (see Sinsheimer PDF).
Thymine Dimer Formation:
Determine the absorption spectrum (230-300 nm) of thymine (25 µg/ml). Place 250 µl of a solution of thymine (500 µg/ml) into each of several 10-ml beakers, freeze (-20oC), and irradiate (254 nm) in the freezer (you must start the lamp while at room temperature, and then put it in the freezer). Take samples at various times, thaw, and quantitatively transfer to a 5-ml volumetric flask. Determine the absorbance at 260 nm; when it has dropped to about 0.3, determine the absorption spectrum of the sample to confirm the loss of the characteristic absorption spectrum of thymine. Then irradiate this solution at room temperature, and follow the return of absorbance at 260 nm with time. When the maximum increase in absorption has been achieved, determine a complete absorption spectrum to confirm the formation of thymine. (While frozen, the thymines are juxtaposed, and the photochemical equilibrium between the formation and splitting of dimers favors dimer formation. In solution, however, the dimers are split by the re-irradiation, and then the monomers diffuse away, and cannot be re-dimerized.)
The thymine dimers (after irradiation while frozen) can be chromatographed and separated from residual thymine
(see Smith PDF). While the use of radioactive thymine would greatly facilitate these studies, the chromatogram can be cut into 1-cm strips (crosswise), and eluted in water, and the absorbance checked at 260 nm. The solutions suspected of containing the thymine dimer (see Smith PDF) can be re-irradiated as above to split the dimer and yield thymine, which now will absorb at 260 nm.
UV SURVIVAL CURVES
Strains AB1157 (wild-type), AB1886 (uvrA6), AB2463 (recA13), and AB2480 (uvrA6 recA13) can be obtained from the Coli Genetic Stock Center, Department of Human Genetics, Yale University School of Medicine, New Haven, CT 06510. This experiment requires some expertise in microbiological techniques, and a calibrated UV lamp at 254 nm. One can roughly calibrate the lamp by exposing the uvrA6 strain to UV radiation for different times, and comparing the survival with the data shown in Figure 1.
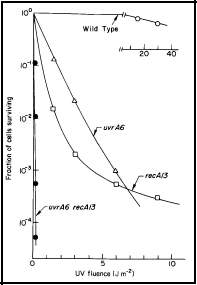
Figure 1. UV radiation survival curves for DNA repair deficient mutants of E. coli K-12. The uvrA6 mutation blocks nucleotide excision repair, and the recA13 mutation blocks recombinational DNA repair. Note that the double mutant, uvrA6 recA13 is very much more sensitive to UV radiation than either of the single mutants, indicating that the two single mutants are involved in separate pathways of DNA repair. Since the two single mutants have about the same sensitivity, it indicates that nucleotide excision repair and recombinational DNA repair are of about equal importance to the survival of UV irradiated E. coli. [Modified from Howard-Flanders and Boyce, 1966] [see Basic Ultraviolet Radiation Photobiology]
The survival curves in Figure 1 can be approximated as follows: The cells are harvested after overnight growth in yeast extract-nutrient broth, and diluted 1:10 in buffer to give ~1 x 108 cells /ml. About 10 ml of cells in a 10-cm Petri dish on a shaker platform are UV-irradiated (8-W GE germicidal lamp at ~47 cm above the cells, giving a fluence rate of ~1 Jm-2 s-1) with various fluences, and appropriately diluted (estimated from Figure 1) to yield about 200 survivors per plate when 0.1 ml is spread on a 10-cm Petri plate. Colonies may be counted after 24-48 h of growth, and the cell surviving fractions determined.
PHOTOREACTIVATION
Spread about 2 x 108 cells of strain AB2480 (uvrA6 recA13) per plate. Prepare 6 plates. Immediately UV-irradiate 4 of the plates without their lids with 0.4 Jm-2 (survival is ~7 x 10-6). Place a Pyrex Petri dish lid full of water, as a UV and infrared filter, on top of 3 of the UV-irradiated plates, and on one non-UV-irradiated plate (without their plastic lids). Place two daylight-fluorescent bulbs 5 cm above the agar surface of these plates, and irradiate the UV-irradiated plates for 1, 5, and 10 min, and the non-UV-irradiated plate for 10 min. Incubate all plates at 37oC for 24-48 hours, and determine the effect of photoreactivation on survival. [see module on Photoreactivation]
05/03/10
03/29/13
03/20/14