PHOTOREACTIVATION
John Jagger
110 Barnett Blvd. #105
Highland Village TX 75077
Deceased 12/27/14 [Obituary]
1. Introduction
Photoreactivation (PR) is the recovery from biological damage caused by UV-C radiation (180-290 nm) or UV-B radiation (290-320 nm) by simultaneous or subsequent treatment with light of longer wavelength (PR light). Early writers often used the older terminology of far UV (<300 nm) and near UV (300-380 nm).
The photoreactivation of viruses and cells has been observed both for killing and mutation, as well as some other effects. It has been found in all taxonomic classes, including viruses and transforming principles, as well as nonplacental mammals. There are exceptions in all classes; it is not found in placental mammals, including humans.
Excellent recent reviews include G.B. Sancar (2000), especially on PR enzymes of Escherichia coli and the yeast Saccharomyces cerevisiae, and Beukers et al. (2008), especially on the PR enzyme of the blue-green alga Anacystis nidulans.
2. Early History
The older literature was reviewed by Dulbecco (1955) and by Jagger (1958, 1967).
The first clear evidence for photoreactivation was provided by Hausser and von Oehmcke (1933), who demonstrated that UV-C exposure strongly darkened banana skin, but the effect was reversed by subsequent irradiation with the mercury arc lines at 366, 405, and 435 nm. This, in fact, was the first action spectrum for photoreactivation. The authors did not explore the phenomenon.
Kelner (1949a) was the first to recognize the importance of PR, when he illuminated spores of the bacterium Streptomyces griseus with PR light after irradiation with light from a germicidal lamp (254 nm). He subsequently found PR of killing in Escherichia coli B/r, and in the fungi Penicillium notatum and Saccharomyces cerevisiae, as well as PR of mutation to phage resistance in E. coli (Kelner, 1949b). Shortly thereafter, Dulbecco (1950) found PR of killing of T2 bacteriophage in E. coli B. In that and later work (see Dulbecco, 1955), he determined that the phage can be irradiated with 254 nm UV when still outside the host bacterium, but PR light is effective only after phage infection, that there is a dark reaction(s) sensitive to temperature, and PR occurs for all the T phages.
Kelner (1949b) compared the UV-C dose for a given survival (dark survival dose) to the UV-C dose that will give the same survival after maximum PR (light survival dose), and found it to be constant. In other words, PR behaves as if it simply reduces the UV-C dose, and he called this the "dose-reduction principle". He found this dose-reduction factor to be about 0.35 for survival of E. coli B/r (Figure 1). We more commonly use the term photoreactivable sector (1- dose-reduction factor), which would therefore be 0.65 for B/r, meaning that about 65% of the UV-C damage is eliminated by maximal PR treatment. Similar results were found by Novick and Szilard (1949) for B/r, and also for mutations to resistance of phage T1, T4, and T6 in B/r, who concluded that whatever chemical effect produces PR of survival of the bacterium is the same as that which produces mutation. Photoreactivable sectors range from low values in the 0.2 range for some phages to 0.65 in E. coli B/r, through 0.82 for S. griseus, to a maximum of 1.0 in Halobacterium cutirubrum (Eker et al., 1991), which means that all of the UV-C lethal damage in this extreme halophilic archaebacterium can be repaired by photoreactivation.
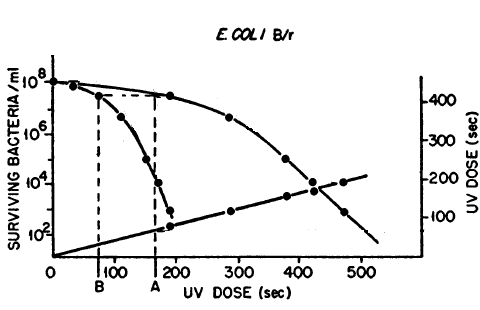
Figure 1. Dose-reduction factor. Survival of E. coli B/r after UV-C dose (left curve) and after UV + maximum PR (right curve) plotted against UV dose. If the UV dose with PR (A) is plotted against the UV dose for the same survival without PR (B), a straight line is obtained that passes through the origin, showing that optimal PR treatment produces a constant reduction of the effects of the UV dose at all survivals (modified from Novick & Szilard, 1949).
Kelner (1951) obtained action spectra for PR of killing in both E. coli B/r and S. griseus. The former showed a peak at 375 nm and no effect above 476 nm, while S. griseus showed a peak at 435 nm, with no effect above 494 nm; the peak at 435 nm suggested porphyrin as a possible chromophore for PR in S. griseus. Dulbecco (1950) obtained an action spectrum for PR of killing of T2 phage in E. coli, in which the range was 313-436 nm, with a peak at 366 nm. These PR action spectra involved wavelengths longer than the known absorption of T2, and therefore showed that the chromophore for PR must be in the bacterium and not in the phage (this was before the Hershey-Chase 1952 experiments showed that only phage DNA enters the host cell). He later suggested (Dulbecco, 1955) that PR in E. coli and Streptomyces could both be produced by a flavin chromophore.
Jagger and Latarjet (1956) obtained more precise action spectra for PR of killing in E. coli B/r and phage T2 in B/r (Figure 2), using a quartz double-traverse double-prism monochromator, which had high band discrimination (double prisms) and no rotatory dispersion (double traverse). They used a high-pressure mercury arc source, with several emission lines, but with a continuous background at lower intensity between those lines from which wavelength bands could be selected by the monochromator without interference from adjacent more intense mercury lines. The action spectra for the two systems were closely similar (peaks at 350 and 380 nm), indicating that the chromophore for PR of bacterial killing was the same as for phage killing. The range of effectiveness was 313-475 nm, but with small valleys for E. coli at 334 nm and for E. coli and T2 at 366 nm; further experiments eliminated the possibility that these valleys resulted from the relatively high intensities of the mercury arc emission lines at those wavelengths. The valley at 366nm was later explained in terms of "indirect PR" (see Section 5c).
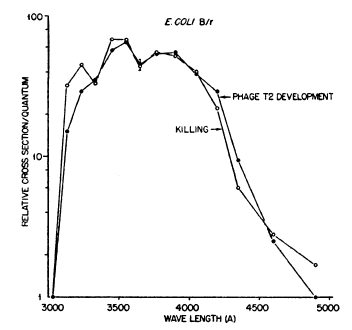
Figure 2. Action spectra for photoreactivation in starved log-phase E. coli B/r and for T2 phage in B/r. The valleys at 334 nm and 366 nm are real (Jagger, 1958, modified from Jagger & Latarjet, 1956).
3. Later In Vitro Studies
Discovery of PR enzyme. The first clear evidence that genes could be made of DNA was demonstrated by the classic finding of Avery, MacLeod, and McCarty (1944) that pneumococcal transforming principle was DNA. Confirmation of this conclusion was the Hershey and Chase (1952) work showing that only the DNA of phage entered the bacterium, proving that phage genes are made of DNA, and not protein.
Once the Hershey-Chase result was known, it could be seen that Dulbecco's 1950 finding of photoreactivation of phage showed that photoreactivable damage could reside in DNA. Final evidence came from the report of Rupert, Goodgal, and Herriott (1958) that transforming DNA of Hemophilus influenzae could be photoreactivated by extracts of either E. coli or baker's yeast (S. cerevisiae). The active principle contained an enzyme-like agent that was called the "PR enzyme".
Further work of Rupert (1962a, b) showed that the PR enzyme combines with UV-C-irradiated DNA, and is thereby stabilized against inactivation by heat and heavy metals. The binding and stabilization are both eliminated by the action of PR light, which leads to DNA repair and enzyme release. This work revealed the general mechanism of photoreactivation, and paved the way for future molecular studies.
Nature of the photoreactivable DNA lesion. All nucleic acid bases absorb UV-C and are damaged by it. The pyrimidines are ten times as sensitive as the purines, and thus are primary sites of damage.
In 1958, Beukers, Ijlstra & Berends made the seminal finding of UV-C production of thymine dimers in a frozen solution of thymine (see also Beukers & Berends, 1960). The dimers were formed by production of a cyclobutane ring involving the 5,6 double bonds of two thymines. This resulted in loss of the 260nm absorption peak. Subsequent irradiation at the same UV-C wavelength of the thawed thymine preparation was found to break the dimer, restoring the original thymines and the 260nm absorption peak (photoreversal). In 1962, Wulff and Rupert showed that more than 90% of thymine dimers are eliminated from transforming DNA by the PR enzyme from yeast. This was the first demonstration that thymine dimers caused biological damage. In DNA the thymine dimer would connect two parallel thymines, lying one above the other, in the same strand of the double helix.
R.B. Setlow and Setlow (1962) and J.K. Setlow & Setlow (1963) showed that transformation by H. influenzae DNA inactivated by large doses of 280 nm radiation could be reactivated by subsequent irradiation at 239 nm, and that the mechanisms involved were the production of thymine dimers at the long wavelength, and their reversal at the short wavelength. This showed that direct photoreversal could occur in a biological system, and provided a model for photoreactivation. Later work (R.B. Setlow et al., 1965; J.K. Setlow et al., 1965) showed that dimers in polynucleotides can be formed between any pyrimidine pair, but are formed at lower rates than thymine dimers; all can be split by PR enzyme. These dimers are now called cyclobutane pyrimidine dimers (CPDs) (Figure 3).
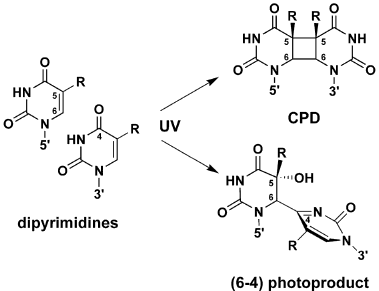
Figure 3. Structures of DNA lesions induced by UV-C radiation. CPD = cyclobutane pyrimidine dimer. R = H or CH3. (From Li et al., 2006).
It had long been known that cells irradiated with UV-C can recover from some of the damage by treatments after irradiation, such as holding the cells in a non-nutrient medium at room temperature (liquid-holding recovery), or by the nature of the plating medium (see Jagger, 1967). In 1963, R.B. Setlow et al. found that the UV-induced blocks to DNA synthesis in E. coli, first shown by Kelner (1953), are permanent in the highly UV-sensitive strain E. coli Bs-1, while in the UV-resistant strain B/r they are only temporary. They found that the recovery of DNA synthesis in the dark does not result from dimer splitting, suggesting that they were physically removed from the DNA. This was the first suggestion of excision dark repair of DNA.
Confirming this idea, R.B. Setlow & Carrier (1964) found that, during the period of dark recovery from UV-C treatment, dimers disappear from the DNA of the radiation-resistant strain B/r of E. coli, but move as oligonucleotides from the acid-insoluble to the acid-soluble fraction of the cells. They are not photoreactivable in the acid-soluble fraction. In the UV-sensitive strain E. coli Bs-1, the dimers remain insoluble and photoreactivable. Similar results in E. coli K-12 strains were reported by Boyce and Howard-Flanders (1964) at nearly the same time. These were the first reports of nucleotide excision repair (NER), also called cut-and-patch repair. It involves removal of short segments of the DNA strand containing the UV-C damage, followed by a patch using the sequence information on the other strand. This finding was of great importance, as it was the beginning of knowledge of dark-repair mechanisms, now known to be important for other types of DNA damage than dimers (see module on Basic Ultraviolet Radiation Photobiology).
Killing and photoreactivation of Streptomyces griseus conidia were measured by Jagger et al. (1967), using vacuum-UV and UV-C radiations (150-270 nm). Photoreactivation dropped to zero at 180 nm, suggesting that CPDs were no longer produced below this wavelength. Preiss and R.B. Setlow (1956) had previously shown that DNA and protein absorption cross-sections were not significantly different at wavelengths below 220 nm.
4. The Photochemical Mechanism
Considering that the PR enzyme breaks carbon-carbon bonds (forming a cyclobutane ring), Minato and Werbin (1972) proposed that it be called a photolyase. Over the years, it has been purified to various degrees from cell extracts. The first of these was by Minato & Werbin (1971), who purified it 70,000-fold from 50 pounds of baker's yeast (S. cerevisiae). In 1980, Iwatsuki et al. concluded that reduced flavin adenine dinucleotide (FADH) is a chromophore in yeast photolyase.
Schild et al. (1984) cloned the gene, PHR1, of S. cerevisiae and determined its map position. They also isolated a plasmid containing a DNA insert that was shown to restore PR in a PHR1 strain. They showed that the plasmid contains the gene PHR1 rather than a suppressor of the PHR1 mutation.
In later work, using recombinant DNA techniques, G.B. Sancar et al. (1987) obtained mg quantities of the yeast photolyase PHR1 at >95% purity . This was produced in E. coli cells by a plasmid containing the PHR1 gene of S. cerevisiae. This nearly pure photolyase permitted molecular characterization of the photolyase chromophore(s). The protein showed an absorption spectrum with a peak at 377 nm and valley at ~320 nm, characteristic of 1,5-reduced FAD. Boiling (to denature the protein) and centrifugation then revealed in the supernatant a typical oxidized flavin with an additional lower absorbance peak at 450 nm. Further studies showed that the E. coli photolyase, like that of yeast, contains a single non-covalently bound reduced FAD cofactor.
G.B. Sancar (2000) outlines the development of our knowledge of the photochemical mechanisms. All photolyases studied to date are monomeric proteins of 55-65 kDa molecular weight, and contain two chromophores. One is the antenna molecule, and the other is always the reduced FAD anion (FADH-), which is the active-site cofactor of the enzyme. One antenna chromophore is 5,10-methenyl tetrahydrofolate (MTHF), found in the bacteria E. coli and Bacillus firmus, and the fungi Saccharomyces cerevisiae and Neurospora crassa (the "folate class"). Another antenna chromophore is 8-hydroxy-5-deazariboflavin (8-HDF), expressed in the archaebacterium Methanobacterium thermoautotrophicum, the actinobacterium Streptomyces griseus, the cyanobacterium Anacystis nidulans, and the alga Scenedesmus acutus (the "flavin class").
Aziz Sancar (1994, 1996) and Kao et al. (2005) describe the energy transfers involved in the actions of chromophores for PR (Figure 4). PR light is absorbed by an antenna molecule, which then transfers its excitation energy to FADH-, forming excited FADH-*. The excited flavin lies adjacent to the pyrimidine dimer to which it transfers an electron, breaking the cyclobutane ring of the dimer, and leaving the flavin as a ground-state reduced radical (FADH*). Electronic rearrangement restores the DNA bases to normal, in the process transferring an electron to the reduced flavin radical, restoring the active-site flavin cofactor (FADH-). It is important to note that PR light can be absorbed by an antenna molecule and/or by the active-site cofactor FADH- (G.B. Sancar et al., 1987).
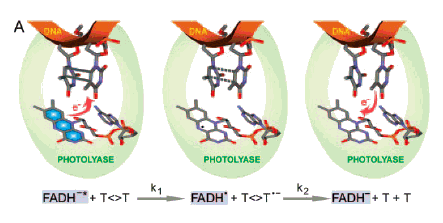
Figure 4. Schematic representation of repair of thymine dimers by CPD photolyase. The key catalytic reactions are given at the bottom. Left: FADH-, the active-site cofactor of the photolyase, excited by a PR photon (blue, FADH-*), transfers an electron to the dimer (T<>T), which has been flipped out of the DNA double helix (red) by the photolyase. Center: The electron transfer breaks the cyclobutane ring of the dimer, leaving 1) a ground-state reduced flavin radical (FADH*), 2) a thymine, and 3) a reduced thymine. Right: Electronic rearrangement restores both thymines to normal, the electron being transferred back to the ground-state reduced flavin radical (FADH*), restoring the original active-site cofactor (FADH-). (Kao et al., 2005).
In 1995, Park et al. determined the 3-dimensional crystallographic structure of the E. coli CPD photolyase to a resolution of 2.3 Å. [See G.B. Sancar (2000) for color photo of the entire E. coli photolyase, and the DNA binding domain of yeast photolyase.] This and work by A. Sancar (1994, 1996) and by Van de Berg and G.B. Sancar (1998) revealed that the photolyase flips the pyrimidine dimer out of the DNA double helix to fit into a hole in the active site of the protein (Figure 4). The dimer lacks the normal hydrogen bonding of the bases with their partners on the other DNA strand and, having lost its base aromaticity, has lower stacking interactions with adjacent bases in the same strand, thus permitting relatively easy rotation of the dimer and its ribose sugars around the single bonds of the phosphate backbone, and out of the double helix.
The photolyase of E. coli repairs relaxed and supercoiled DNA with equal efficiency (G.B. Sancar et al., 1985). The rate of repair of DNA in cells by photolyase is much slower in regions containing nucleosomes. It is also slower in a transcriptionally active gene, apparently being blocked by RNA polymerase (G.B. Sancar, 2000). Action spectrum analysis by Payne and Sancar (1990) showed that single and double-stranded DNA are repaired by the E. coli photolyase with the same overall quantum yield, and that FADH- can serve as a chromophore with almost equal efficiency as MTHF, the action spectrum peak at 380 nm (Figure 5) reflecting absorption by both MTHF and FADH- chromophores.
The excitation energy transfer from MTHF to FADH- occurs by radiationless Förster resonance over the 17 Å that separates the two molecules, with an efficiency of about 70%. The flavin-class antenna molecule, 8-HDF, is about the same distance away, but is better aligned with FADH-, showing a 98% efficiency of Förster transfer (Beukers et al., 2008, A. Sancar, 2008).
Photolyases have now been purified from the bacteria E. coli, S. griseus, and Anacystis nidulans, the archaebacterium M. thermoautotrophicum, the yeast S. cerevisiae, and the alga S. acutus. Photolyase genes have been cloned and sequenced for three of these organisms, as well as from the archaebacterium Halobacterium halobium (see G.B. Sancar, 1990). See Figure 5 for action spectra.
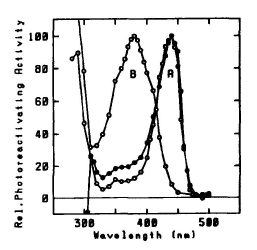
Figure 5. Action spectra for photoreactivation.
A: Closed circles: Halobacterium cutirubrum in vivo
(max = 440 nm). Open circles: photolyase of Anacystis nidulans (flavin class) (
max = 440 nm). B: photolyase of Saccharomyces cerevisiae (folate class) (
max = 380 nm). Maxima normalized to 100. (Eker et al., 1991)
A broad review of photoreactivation, especially of the work of researchers in the Delft Technological University in The Netherlands, was published by Beukers et al. in 2008. This includes detailed crystal studies of the CPD photolyase (flavin class) of Anacystis nidulans, showing close parallels with the structure of the E. coli photolyase (folate class). They also show a remarkable visualization by atomic force spectroscopy of the E. coli photolyase bound to 830-bp ds-DNA fragments, which suggests that the photolyase slides along DNA until it finds a 30o kink containing the CPD.
5. Other Aspects
a. Repair of (6-4) photoproduct. The major UV-C photoproduct in DNA is the cyclobutane pyrimidine dimer (CPD), which typically accounts for about 75% of UV-C photoproducts in DNA. The second most common DNA photoproduct is 6-4'-pyrimidin-2'-one pyrimidine, called the (6-4) photoproduct (Figure 3), discovered by Varghese and Wang (1967) in Streptomyces strains, which typically accounts for about 25% of UV-C photoproducts (see Kim et al., 1994). The (6-4) photoproduct has an absorption maximum at ~320 nm (Figure 6A).
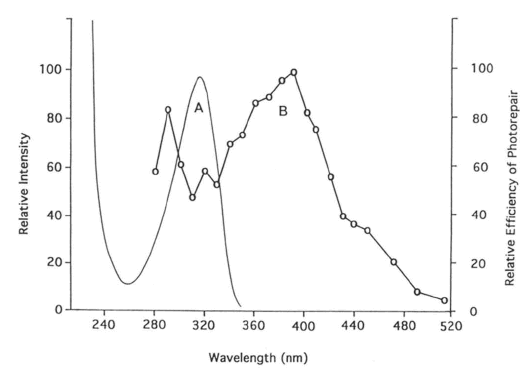
Figure 6. (A) Absorption spectrum of T(6-4)T in duplex DNA. (B) Action spectrum of D. melanogaster (6-4) photolyase repairing this photoproduct, showing a peak at 290 nm, a valley at ~320 nm, and a peak at 400 nm, which falls between the peaks for flavin and folate class photolyases (see Figure 5). (From Kim et al., 1994)
High doses of light at 313 nm convert the T(6-4)T photoproduct to its Dewar isomer, which is subject to nucleotide excision repair (NER), but not photolyase repair. Under sunlight conditions, various amounts of the UV-C photoproduct should be in the Dewar form.
In 1993, Todo et al. discovered a (6-4) photolyase in Drosophila melanogaster. Kim et al. (1994) showed that the (6-4) photoproduct, but not its Dewar isomer, is the substrate for this enzyme, that the efficiency of repair per incident photon is very low compared with CPD photolyases, and that the action spectrum has a maximum at 400 nm and a minimum around 320 nm (Figure 6). This action spectrum also reflects the later finding by Todo et al. (1996) that a gene for Drosophila (6-4) photolyase codes both a folate chromophore (maximum 380 nm) and a flavin chromophore (maximum 440 nm) (see Figure 5).
Like the CPD photolyases of both the folate class and the flavin class, the (6-4) photolyase of D. melanogaster restores the original pyrimidines after repair. It appears to repair via the same unstable oxetane ring intermediate produced during formation of the (6-4) photoproduct (Kim et al., 1994). The (6-4) photolyase does not bind to CPDs. In 2001, Hitomi et al., working with the (6-4) photolyase of the frog Xenopus laevis, showed that two histidines of the protein chain, not present in CPD photolyases, are essential to the (6-4) photolyase mechanism, perhaps stabilizing formation of the oxetane intermediate.
(6-4) Photolyases have been found so far only in some higher eukaryotes, and complementary DNAs have been cloned from D. melanogaster, X. laevis, Danio rerio and A. thaliana (see Hitomi et al., 2001). Xiphophorus signum (platyfish) shows (6-4) photoproduct induction by UV-C at a much lower frequency than CPD induction, but PR of CPD was rapid and twice the rate of PR of (6-4) photoproduct (Meador et al., 2000).
Jagger et al. (1970) determined an action spectrum for PR of killing in Streptomyces griseus that showed the major peak at 436 nm originally observed by Kelner, and a new peak at 313 nm (for review, see Jagger, 2004). Both S. coelicolor and a PHR1 mutant of S. griseus also showed the 313 nm peak, but no PR above 405 nm. The 313 nm PR peak in all three strains showed little or no dependence upon temperature or dose rate of the PR light. Later work showed rapid loss of the (6-4) photoproduct at 313 nm in all three strains, but with no effect on killing in the wild type (see Ikenaga et al., 1971). In S. coelicolor and S. griseus PHR1, which appear to lack CPD photolyase, these 313 nm effects appeared to be direct photochemical action on the (6-4) photoproduct. They called this new type of direct photoreversal Type III PR. Supportive of this interpretation is that (6-4) photolyase has not been found in prokaryotes (Hitomi et al., 2001).
b. Cytoplasmic photoreactivation. Some studies of cytoplasmic PR have been negative. For example, von Borstel & Wolff (1955) found no PR of hatchability of eggs of the wasp Habrobracon after cytoplasmic irradiation with UV-C. The eggs were irradiated when the nucleus was close to one surface of the egg, so that one could irradiate either the nuclear side or, by turning the egg over, the cytoplasmic side, when the nucleus would be shielded by cytoplasm.
However, Jagger et al. (1969) irradiated living cells of Amoeba proteus with a microbeam of UV-C radiation followed by PR light. The amebae were flattened under a cover slip, so that either the nucleus or the cytoplasm could be irradiated with little overlap, thus eliminating shielding of the nucleus by cytoplasm. Killing by UV-C of flattened amebae results equally from damage to the nucleus and to the cytoplasm, while division delay results almost entirely from cytoplasmic damage. In unflattened amebae, both effects are due to cytoplasmic damage, owing to cytoplasmic shielding of the nucleus. Both killing and division delay showed significant PR after nuclear irradiation, but a much larger PR of both effects after cytoplasmic irradiation. This was a clear demonstration of cytoplasmic PR, presumably due largely to effects on the DNA of mitochondria, which were later shown to have photolyase (Green & MacQuillan, 1980). In the yeast, S. cerevisiae, Prakash (1975) found that PR treatment resulted in a decrease of dimers from both nuclear and mitochondrial DNA. Mitochondrial lysates of Xenopus laevis oocytes show both photoreactivation and excision repair of UV-C-irradiated DNA (Ryoji et al., 1996).
The cytoplasmic PR could also be due partly to action on transfer RNA or ribosomal RNA. The cytoplasm of A. proteus contains much more RNA than DNA. PR of RNA has not been studied very much, either in vivo or in vitro. Uracil and cytosine dimers are split by yeast photolyase, but with lower efficiency than thymine dimers (see Section 3). DNA photolyases work on RNA as well as on DNA (Kim & Sancar (1993), and single-stranded DNA is highly photoreactivable (Payne and Sancar (1990). The finding that thiouridines in some bacterial tRNAs are chromophores and targets for growth delay induced by wavelengths as low as 310 nm (Section 5c) shows that tRNA can be a UV-B target.
There are few studies of PR in plant viruses, most of which contain single-stranded RNA. Tobacco mosaic virus is not photoreactivable when intact, but its RNA does show inactivation and PR when separated from the protein coat, in which form it is five times as sensitive to UV-C as the intact virus (see Jagger, 1967). Bawden & Kleczkowski (1952) observed a small PR of the spherical tomato bushy stunt virus in leaves of Nicotiana glutinosa and tobacco necrosis virus in French bean (Phaseolus vulgaris).
c. Photoprotection and indirect photoreactivation. The phenomenon of photoprotection, in which longer wavelengths of light given before exposure to UV-C can result in higher survival, was discovered by Weatherwax (1956) in E. coli. (This is not to be confused with the same term in photomedicine, which is a chemical protection against radiation.) Jagger (1960) showed that the photoprotection rate in E. coli B has only a slight dependence upon temperature, and no dependence upon dose rate of the PR light, indicating that it is different from enzymatic PR. The effective wavelengths lie in the range 310-400 nm, with a single peak at 340 nm (Jagger & Stafford, 1962).
It had long been known that UV-A radiation (320-400 nm) could induce a growth delay in E. coli (Hollaender, 1943). The action spectrum for such growth delay in E. coli B was found by Jagger et al. (1964) to be identical to the action spectrum for photoprotection, suggesting that photoprotection operates by inducing a growth delay. Such a delay could provide more time for dark repair, as indicated by the finding of complete overlap of photoreactivation and liquid-holding recovery in E. coli B (Castellani et al., 1964).
Ramabhadran and Jagger (1975) later showed that UV-A-induced growth delay was not photoreactivable, and not affected by dark-repair systems. This showed that DNA damage was not involved.
In 1969, Favre et al. showed that 334 nm irradiation of E. coli tRNA produces an adduct between 4-thiouridine, an unusual nucleotide found in some bacterial tRNAs, and a nearby, but not adjacent, cytidine residue in the tRNA. This 4-thiouridine-cytidine adduct would prevent amino acid charging of the tRNA, and thus reduce protein synthesis. Ramabhadran (1975) showed that the absorption spectrum of E. coli valyl tRNA, which contains 4-thiouridine, matched very closely the action spectra for growth delay and for inhibition of net RNA synthesis, and concluded that the chromophore and target for UV-A-induced growth delay in actively growing E. coli was likely 4-thiouridine in transfer RNA. This was confirmed by Thomas and Favre (1975) in E. coli K-12, who showed that the amount of cross-linked tRNA is closely correlated to growth delay, and by Ramabhadran and Jagger (1976), who showed that UV-A irradiation of E. coli partially inactivates certain tRNA species. This is interpreted by the cell in a manner similar to that of amino-acid starvation, causing a temporary rise in levels of guanosine tetraphosphate (ppGpp) and shut-off of net RNA synthesis, resulting in growth delay. Tsai and Jagger (1981) found that mutants lacking 4-thiouridine showed no photoprotection. Since growth delay is responsible for photoprotection, it follows that a chromophore and target for both UV-A-induced growth delay and photoprotection in E. coli is 4-thiouridine in tRNA.
Later work by Favre and coworkers (Thomas et al., 1981; Thiam and Favre, 1984) showed a short UV-A-induced growth lag in E. coli nuv-, which lacks 4-thiouridine. However, this strain shows a considerable increase in ppGpp. It has another tRNA chromophore, 5-methylaminomethyl-2-thiouridine, induced by UV-A wavelengths
<350 nm, and present in the anticodon loops of some tRNAs.
Thus, both 4-thiouridine and 2-thiouridine in tRNAs are chromophores for photoprotection and growth delay in E. coli.
Jagger and Stafford (1965) found that a PR-deficient strain of E. coli B phr- shows PR at 334 nm but not at 405 nm, and exhibits dose rate and temperature independence. This PR is therefore indistinguishable from photoprotection and growth delay. Since PR is defined as "recovery from biological damage caused by UV-C radiation by simultaneous or subsequent treatment with light of longer wavelength", they called this indirect photoreactivation (later Type-II PR). Thus, indirect PR is non-enzymatic and due to induction of a growth delay that permits more time for dark repair.
Later work (Jagger et al., 1969) showed that all of the PR in E. coli B phr-, and part of the PR in strain B, does not involve thymine-dimer splitting, and therefore does not use photolyase. Their action spectra (for review, see Jagger, 2004) showed that PR at wavelengths below 366 nm in starved log-phase E. coli B/r could be explained as 20% direct PR (enzymatic) and 80% indirect PR (non-enzymatic). This explained the 366nm valley in the action spectrum of Jagger and Latarjet (Figure 2) for PR in starved log-phase E. coli B/r, where indirect PR (peaking at 340 nm) accounts for most of the PR at wavelengths below 366 nm, while photolyase PR explains most of the PR above 366 nm. It did not, however, explain the 334 nm valley of Jagger & Latarjet (1956).
Consistent with this, they obtained an action spectrum for PR of stationary-phase E. coli Bs-1 (deficient in dark repair) showing a single maximum at 380 nm (see Jagger, 2004). It contains neither of the valleys found by Jagger and Latarjet (Figure 2) for log-phase E. coli B/r
at 334 nm and 366 nm. Since strain Bs-1 shows no dark repair, it should also show no indirect PR, so PR in this organism should reflect the true action spectrum of the E. coli CPD photolyase (folate class), which is consistent with later action spectra (see Figure 5).
Photoprotection is less widespread than photoreactivation. It was found in E. coli, Pseudomonas aeruginosa, Staphylococcus aureus, and the protozoa Amoeba proteus and Colpidium colpoda, but not in Streptomyces griseus and Saccharomyces cerevisiae (Jagger & Stafford, 1962).
Photoprotection and indirect PR are probably only important in systems that do not already have a built-in growth delay (allowing more time for dark repair), such as by being starved or in the stationary growth phase. The negative findings reported in 1962 by Jagger and Stafford might have been positive had more attention been paid to growth conditions, the role of growth delay in photoprotection being shown only later in 1964.
While CPDs and (6-4) photoproducts can account for much of the PR in many cells, effects like indirect PR can complicate the picture, as illustrated by the later analysis (outlined above) of the original E. coli PR action spectrum of Jagger and Latarjet.
d. Biological range of photoreactivation. Biological ranges of PR have been reported in several sources, including Jagger (1958, 1967), Cook & McGrath (1967), A. Sancar (1994), and Goosen & Moolenaar (2008).
CPD photolyase has been found in a wide variety of bacteria and archaea, but there are notable exceptions. It has not been found in Bacillus subtilis or Hemophilus influenzae, or in the archaeon Methanococcus vannielii. It is found in the fungi Neurospora crassa and Saccharomyces cerevisiae, but not in Schizosaccharomyces pombe.
Goosen & Moolenaar (2008) conducted an extensive study of UV damage repair in bacteria. They determined the complete genome sequences of 519 species of eubacteria and 44 species of archaebacteria, and analyzed the evolutionary development of photolyases, CPD-DNA glycosylases, nucleotide excision repair (NER) proteins, and UV-damage endonucleases (UVDE), first identified (as SPDE) by Bowman et al. (1994) in Schizosaccharomyces pombe. The UVDE protein recognizes both CPDs and (6-4) photoproducts, as well as several other non-UV-induced adducts; it occurs in B. subtilis and in the highly radiation-resistant Deinococcus radiodurans. Its presence in S. pombe and B. subtilis might explain their lack of individual CPD and 6-4 photolyases.
Only 25% of the archaea analyzed contain a photolyase gene. Four species of archaea contain UVDE, but only one (Sulfolobus acidocaldarius) has both UVDE and photolyase. All species in the order Halobacteriales have more than one photolyase homolog, and all three of the NER excision genes, and a few like Haloarcula marismortui contain all four types of the repair enzyme listed above, properties perhaps essential for organisms living under extreme conditions of heat and salinity. Not surprisingly, fewer deep-sea species have photolyase.
Goosen & Moolinaar (2008) note that photolyases were the first repair enzymes, and evolved very early in evolution, since the early prokaryotes would need protection against solar UV before stratospheric ozone developed. Homologs of CPD photolyase are found in only about 50% of the eubacteria studied, suggesting gene loss, which could have happened after their invasion of eukaryotes, where many would no longer be exposed to UV-A and UV-B (although E. coli has photolyase). They also suggest that, since 50% of bacteria that have UvrABC excision proteins for dark repair also have CPD photolyase, the NER proteins may have evolved primarily for the repair of (6-4) photoproducts, and other types of damage not induced by UV. They suggest that cryptochromes (flavoproteins involved in control functions) evolved from photolyases, since cryptochromes occur only rarely in prokaryotes.
CPD photoreactivation is found in the protozoa Amoeba proteus and Colpidium colpoda, and in the plants Arabidopsis thaliana and Scenedesmus acutus (alga). It is found in a wide variety of animals, although often only in certain tissues. It is not found in the nematode Caenorhabditis elegans. It has been found in non-placental mammals, including the rat kangaroo Potorous tridactylus (Harm, 1978) and the opossum Monodelphus domestica (Ley, 1984).
Despite early reports to the contrary, current evidence indicates that humans do not have DNA photolyase. Li et al. (1993) found that, while CPD photolyase was easily detectable in cells of E. coli and yeast, and in rattlesnake cell-free extracts, none was detected in cell-free extracts from HeLa cells or human white blood cells, using an assay capable of detecting 10 molecules of photolyase per cell. And Chigancas et al. (2000) found that HeLa cells showed PR only after introduction of a photolyase gene from Potorous tridactylus, thus demonstrating that their experimental conditions for PR treatment of HeLa cells would permit PR if a photolyase were present. Finally, sequencing of the human genome is now complete, and all sequences that appear to code for photolyase-like proteins are accounted for by the cryptochromes.
Pyrimidine dimers induced by UV (280-400 nm, peak at 313 nm) in the epidermis of Monodelphis domestica, a small South American opossum, were removed by treatment with PR light (Ley, 1984). Such treatment also resulted in photoreactivation of erythema (Ley, 1985).
Hart et al. (1977) reported that thyroid tumors that developed from transplanted UV-C-irradiated thyroid cells were highly photoreactivable in the small tropical fish Poecilia formosa showing that pyrimidine dimers can induce tumors. R.B. Setlow et al. (1989) later found that PR treatment of a platyfish-swordtail hybrid (Xiphophorus) reduced the incidence of UV-B-induced melanoma to background levels, and Ley et al. (1989) found PR of melanoma induction in the skin of M. domestica. These results showed that CPDs produced by either UV-B or UV-C can act as complete carcinogens for melanoma induction. Cook & McGrath (1967) had earlier failed to find PR activity in skin of the mouse or rabbit.
6. Retrospect
In 1944, physicist Erwin Schrödinger wrote the seminal book, What is Life? (Cambridge University Press) in which he proposed that a complex molecule could contain the genetic code for living organisms. In the early 20th century, Thomas Hunt Morgan had shown that Drosophila genes are arranged in a linear sequence within chromosomes. Also, it was well known that genes showed remarkable stability, a necessity for evolution. Therefore, Schrödinger reasoned that the genetic material had to be a linear molecule of great length and great stability, since such stability could only be attained in what he called "an aperiodic crystal". This was a great insight, and gave hope that the gene might someday be understood in terms of chemistry and physics.
Action spectra for bacterial killing (Gates, 1930) and for killing and mutation of fungal spores (Emmons & Hollaender, 1939) showed peaks around 265 nm, suggesting that nucleic acid was the chromophore for UV biological action. They were cautious in their interpretations; neither claimed that the work showed that nucleic acid was the genetic material of cells. Thus, Gates (1934) hedged his bets by stating that his action spectra for killing of Staphylococcus aureus and its phage was similar to curves "...for the specific absorption of ultraviolet light by protoplasm, by proteins, by certain amino acids and nucleoproteins, and by certain enzymes." And Zelle and Hollaender (1955), concluded (p. 415) that a "growing body of evidence indicates that a major proportion of the effects of ...ultraviolet... radiation on bacteria are indirect and involve a largely unknown chain of reactions occurring between the initial...quantum absorption and the final lethal or mutagenic change."
Thus, Avery et al. (1944) and Hershey and Chase (1952) are the true discoverers of DNA as the genetic material. Even then, it was a conclusion reluctantly accepted. Some workers stuck to the idea that protein, because of its high information content (20 amino acids) was the more likely genetic material, with DNA (only 4 nucleotides) acting as a supporting scaffold. Doubters claimed that a small amount of protein was present in the transforming principle of Avery et al., (1944), and could have carried the genetic information.
However, even though DNA became generally accepted as the genetic material by 1952, the idea of great stability required of the genome caused scientists in those days to assume that DNA could be changed only by mutation by energetic events, such as ionization by X rays or chemical change by UV. The idea that DNA might be constantly damaged by things as ordinary as cellular metabolism would require that the damage be frequently and accurately repaired, a notion at that time inconceivable. No molecular systems were known that could repair a damaged molecule.
PR was discovered for killing and mutation of bacteria by Albert Kelner (1949a). In the next year, Dulbecco (1950) found that T phages could be photoreactivated inside bacteria, pointing to DNA as the target molecule for PR. Yet it is not evident that anyone thought that PR could involve simple DNA repair by an enzyme. Many early studies assumed that it must act by neutralizing poisons in the medium that damaged the DNA (e.g., Novick and Szilard, 1949).
All this changed with the report of Rupert et al. (1958) that PR could be produced in vitro by an enzymatic reaction. It suddenly became evident that DNA could be directly repaired, and that the repair was accurate. Around this time we were becoming aware of the fantastic capabilities of DNA polymerases and the complexity of ribosomes. As the structures of photolyases became known in the 1990s, the abilities of these enzymes to perform such surprising maneuvers as flipping dimers out of the DNA double helix, resonance transfer of energy from distant chromophores, followed by redox repair of dimers, is reminiscent of the complex abilities of photosynthetic reaction centers and the enzymes of the electron-transport systems of oxidative metabolism. Some UV-C repair molecules, such as UVDE, can even repair more than one UV-C photoproduct, as well as several non-UV-induced adducts.
Many early workers in photobiology were motivated by a philosophical consideration: Since life evolved in a world of light, there must be many interactions of biological systems with light, including accommodations for its deleterious effects. Humankind has known for centuries that sunlight provides energy for plant life, and we know that, in the last analysis, since all animals depend upon plants somewhere in the food chain, photosynthesis is a fundamental energy requirement for life on Earth. We now know that the photosynthetic structures of plants are remarkably complex, with many chromophores feeding into a photosynthetic reaction center (hence our beautiful fall colors), and that photosynthesis can operate with basic cofactors other than chlorophyll.
Light also provides control of biochemical processes, as in phototropism and photoperiodism. It is now evident that some control systems utilize cryptochromes in blue-light responses of plants, circadian clocks in fruit flies and mice, and perhaps magnetoreceptors in migratory animals. Cryptochromes bear a molecular similarity to DNA photolyases, which perform repair of light damage. All this reflects the great economy in nature of using similar proteins for widely different functions (see A. Sancar, 2008).
Such complex molecular function was not imagined in the early days of photobiology. It has been a paradigm change, and should caution us not to think too simply about the future of molecular photobiology. Much still remains to be learned.
References
Avery, O.T., C.M. MacLeod & M. McCarty 1944 Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus Type III. J. Exptl. Med. 79, 137-158
Bawden, F.C. & A. Kleczkowski 1952 Ultra-violet injury to higher plants counteracted by visible light. Nature 169, 90-91.
Beukers, R. & W. Berends 1960 Isolation and identification of the irradiation product of thymine. Biochim. Biophys. Acta 41, 550-551.
Beukers, R., A.P.M. Eker & P.H.M. Lohman 2008 50 years thymine dimer. DNA Repair 7, 530-543.
Beukers, R., J. Ijlstra & W. Berends 1958 The effect of ultraviolet light on some components of the nucleic acids. II. In rapidly frozen solutions. Rec. Trav. Chim. 77, 729-732.
Bowman, K.K., K. Sidlik, C.A. Smith, J.S. Taylor, P.W. Doetsch & G.A. Freyer 1994 A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res. 22, 3026-3032.
Boyce, R.P. & P. Howard-Flanders 1964 Release of ultraviolet light-induced thymine dimers from DNA. Proc. Natl. Acad. Sci. USA 51, 293-300.
Castellani A., J. Jagger & R.B. Setlow 1964 Overlap of photoreactivation and liquid holding recovery in Escherichia coli B. Science 143, 1170-1171.
Chigancas, V., E.N. Miyaji, A.R. Muotri, J. F. Jacysyn, G.P. Amarente-Mendes, A. Yasui & C.F.M. Menck 2000 Photorepair prevents ultraviolet-induced apoptosis in human cells expressing the marsupial photolyase gene. Cancer Research 60, 2458-2463.
Cook, J.S. & J.R. McGrath 1967 Photoreactivating enzyme activity in metazoa. Proc. Natl. Acad. Sci. U.S. 58, 1359-1365.
Dulbecco, R. 1950 Experiments on photoreactivation of bacteriophages inactivated with ultraviolet radiation. J. Bacteriol. 59, 329-347.
Dulbecco, R. 1955 Photoreactivation. In, A. Hollaender, Ed. Radiation Biology Vol. II: Ultraviolet and Related Radiations (New York: McGraw-Hill) 365-430.
Eker, A.P.M., L. Formenoy & L.E.A. de Wit 1991 Photoreactivation in the extreme halophilic archaebacterium Halobacterium cutirubrum. Photochem. Photobiol. 53, 643-651.
Emmons, C.W. & A. Hollaender 1939 The action of ultraviolet radiation on dermatophytes. II. Mutations induced in cultures of dermatophytes by exposure of spores to monochromatic ultraviolet irradiation. Amer. J. Botany 26, 467-475.
Favre, A., M.Yaniv & A.M. Michelson 1969 The photochemistry of 4-thiouridine in Escherichia coli tRNA1Val. Biochem. Biophys. Res. Commun. 37, 266-271.
Gates, F.L. 1930 A study of the bactericidal action of ultra violet light. III. The absorption of ultra violet light by bacteria. J. Gen. Physiol. 14, 31-42.
Gates, F.L. 1934 Results of irradiating Staphylococcus aureus bacteriophage with monochromatic ultraviolet light. J. Exptl. Med. 60, 179-188.
Goosen, N. & G.F. Moolinaar 2008 Repair of UV damage in bacteria. DNA Repair 7, 353-379.
Green, G. & A.M. MacQuillan 1980 Photorepair of ultraviolet-induced petite mutational damage in Saccharomyces cerevisiae requires the product of the PHR1 gene. J. Bacteriol. 144, 826-829.
Harm. H. 1978 Damage and repair in mammalian cells after exposure to non-ionizing radiations: I. Ultraviolet and visible light irradiation of cells of the rat kangaroo (Potorous tridactylus) and determination of photorepairable damage in vitro. Mutation Research 50, 353-366.
Hart, R.W., R.B. Setlow & A.D. Woodhead 1977 Evidence that pyrimidine dimers in DNA can give rise to tumors. Proc. Natl. Acad. Sci. USA 74, 5574-5578.
Hausser, K.W. & H. von Oehmcke 1933 Lichtbräunung an Fruchtschalen. Strahlentherapie 48, 223-229.
Hershey, A.D. & M. Chase 1952 Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39-56.
Hitomi, K., H. Nakamura, S-T. Kim, T. Mizukoshi, T. Ishikawa, S. Iwai & T. Todo 2001 Role of two histidines in the (6-4) photolyase reaction. J. Biol. Chem. 276, 10103-10109.
Hollaender, A. 1943 Effect of long ultraviolet and short visible radiation (3500-4900 Å) on Escherichia coli. J. Bacteriol. 46, 531-541.
Ikenaga, M., M.H. Patrick & J. Jagger 1971 Photoreactivation of killing in Streptomyces - III. Action spectra for photolysis of pyrimidine dimers and adducts in S. griseus and S. griseus PHR-1. Photochem. Photobiol. 14, 175-187.
Iwatsuki, N., C.D. Joe & H. Werbin 1980 Evidence that deoxyribonucleic acid photolyase from baker's yeast is a flavoprotein. Biochemistry 19, 1172-1176.
Jagger, J. 1958 Photoreactivation. Bacteriol. Rev. 22, 99-142.
Jagger, J. 1960 Photoreactivation. In, A. Hollaender, Ed. Radiation Protection and Recovery (New York, Pergamon Press) 352-377.
Jagger, J. 1967 Introduction to Research in Ultraviolet Photobiology (Old Tappan NJ: Prentice-Hall).
Jagger, J. 2004 Personal reflections on monochromators and action spectra for photoreactivation. J. Photochem. Photobiol. B. Biology 73, 109-114.
Jagger, J. & R. Latarjet 1956 Spectres d'action de la photo-restauration chez E. coli B/r. Ann. Inst. Pasteur 91, 858-873.
Jagger, J. & R.S. Stafford 1962 Biological and physical ranges of photoprotection from ultraviolet damage in microorganisms. Photochem. Photobiol. 1, 245-257.
Jagger, J. & R.S. Stafford 1965 Evidence for two mechanisms of photoreactivation in Escherichia coli B. Biophys. J. 5, 75-88.
Jagger, J., D.M. Prescott & M.E. Gaulden 1969 An ultraviolet microbeam study of the roles of nucleus and cytoplasm in division delay, killing, and photoreactivation of Amoeba proteus. Exptl. Cell Res. 58, 35-54.
Jagger, J., R.S. Stafford & R.J. Mackin, Jr. 1967 Killing and photoreactivation of Streptomyces griseus conidia by vacuum-ultraviolet and far-ultraviolet radiation (1500 to 2700 Å). Radiation Res. 32, 64-92.
Jagger, J., R.S. Stafford & J.M. Snow 1969 Thymine-dimer and action-spectrum evidence for indirect photoreactivation in Escherichia coli. Photochem. Photobiol. 10, 383-395.
Jagger, J., H. Takebe & J.M. Snow 1970 Photoreactivation of killing in Streptomyces: Action spectra and kinetic studies. Photochem. Photobiol. 12, 185-196.
Jagger, J., W.C. Wise & R.S. Stafford 1964 Delay in growth and division induced by near ultraviolet radiation in Escherichia coli B and its role in photoprotection and liquid holding recovery. Photochem. Photobiol. 3, 11-24.
Kao, Y-T., C. Saxena, L. Wang, A. Sancar & D. Zhong 2005 Direct observation of thymine dimer repair in DNA by photolyase. Proc. Natl. Acad. Sci. USA 102, 16128-16132.
Kelner, A. 1949a Effect of visible light on the recovery of Streptomyces griseus conidia from ultraviolet irradiation injury. Proc. Natl. Acad. Sci. USA 35, 73-79.
Kelner, A. 1949b Photoreactivation of ultraviolet-irradiated Escherichia coli, with special reference to the dose-reduction principle and to ultraviolet-induced mutation. J. Bacteriol. 58, 511-522.
Kelner, A. 1951 Action spectra for photoreactivation of ultraviolet-irradiated Escherichia coli and Streptomyces griseus. J. Gen. Physiol. 34, 835-852.
Kelner, A. 1953 Growth, respiration, and nucleic acid synthesis in ultraviolet-irradiated and in photoreactivated Escherichia coli. J. Bacteriol. 65, 252-262.
Kim, S-T. & A. Sancar 1993 Photochemistry, photophysics, and mechanisms of pyrimidine dimer repair by DNA photolyase. Photochem. Photobiol. 57,895-904.
Kim, S-T., K. Malhotra, C.A. Smith, J-S. Taylor & A. Sancar 1994 Characterization of (6-4) photoproduct DNA photolyase. J. Biol. Chem. 269, 8535-8540.
Ley, R.D. 1984 Photorepair of pyrimidine dimers in the epidermis of the marsupial Monodelphis domestica. Photochem. Photobiol. 40, 141-143.
Ley, R.D. 1985 Photoreactivation of UV-induced pyrimidine dimers and erythema in the marsupial Monodelphis domestica. Proc. Natl. Acad. Sci. USA 82, 2409-2411.
Ley, R.D., L.A. Applegate, R.S. Padilla & T.D. Stuart 1989 Ultraviolet radiation-induced malignant melanoma in Monodelphis domestica. Photochem. Photobiol. 50, 1-5.
Li, J., S-T. Kim & A. Sancar 1993 Evidence for lack of DNA photoreactivating enzyme in humans. Proc. Natl. Acad. Sci. USA 90, 4389-4393.
Li, J., T. Uchida, T. Todo & T. Kitagawa 2006 Similarities and differences between cyclobutane pyrimidine photolyase and (6-4) photolyase as revealed by resonance Raman spectroscopy. J. Biol. Chem. 281, 25551-25559.
Meador, J.A., R.B. Walter & D.L. Mitchell 2000 Induction, distribution and repair of UV photodamage in the platyfish Xiphophorus signum. Photochem. Photobiol. 72, 260-266.
Minato, S. & H. Werbin 1971 Spectral properties of the chromophoric material associated with the deoxyribonucleic acid photoreactivating enzyme isolated from baker's yeast. Biochemistry 10, 4503-4508.
Minato. S. & H. Werbin 1972 Excitation and fluorescence spectra of the chromophore associated with the DNA-photoreactivating enzyme from the blue-green alga Anacystis nidulans. Photochem. Photobiol. 15, 97-100.
Novick, A. & L. Szilard 1949 Experiments on light-reactivation of ultra-violet inactivated bacteria. Proc. Natl. Acad. Sci. USA 35, 591-600.
Park, H.W., S-T. Kim, A. Sancar & J. Deisenhofer 1995 Crystal structure of DNA photolyase from Escherichia coli. Science 268, 1866-1872.
Payne, G. & A. Sancar 1990 Absolute action spectrum of E-FADH2-MTHF forms of Escherichia coli DNA photolyase. Biochemistry 29, 7715-7727.
Prakash, L. 1975 Repair of pyrimidine dimers in nuclear and mitochondrial DNA of yeast irradiated with low doses of ultraviolet light. J. Mol. Biol. 98, 781-795.
Preiss, J.W. & R. Setlow 1956 Spectra of some amino acids, peptides, nucleic acids, and protein in the vacuum ultraviolet. J. Chem. Phys. 25, 138-141.
Ramabhadran, T.V. 1975 Effects of near-ultraviolet and violet radiations (313-405 nm) on DNA, RNA, and protein synthesis in E. coli B/r: Implications for growth delay. Photochem. Photobiol. 22, 117-123.
Ramabhadran, T.V. & J. Jagger 1975 Evidence against DNA as the target for 334 nm-induced growth delay in Escherichia coli. Photochem. Photobiol. 21, 227-233.
Ramabhadran, T.V. & J. Jagger 1976 Mechanism of growth delay induced in Escherichia coli by near ultraviolet radiation. Proc. Natl. Acad. Sci. USA 73, 59-63.
Rupert, C.S., S.H. Goodgal & R.M. Herriott 1958 Photoreactivation in vitro of ultraviolet inactivated Hemophilus influenzae transforming factor. J. Gen. Physiol. 41, 451-471.
Rupert, C.S. 1962a Photoenzymatic repair of ultraviolet damage in DNA. I. Kinetics of the reaction. J. Gen. Physiol. 45, 703-724.
Rupert, C.S. 1962b Photoenzymatic repair of ultraviolet damage in DNA. II. Formation of an enzyme-substrate complex. J. Gen. Physiol. 45, 725-741.
Ryoji, M., H. Katayama, H. Fusamae, A. Matsuda. F. Sakai & H. Utano 1996 Repair of DNA damage in a mitochondrial lysate of Xenopus laevis oocytes. Nucleic Acids Res. 24, 4057-4062.
Sancar, G.B. 1990 DNA photolyases: Physical properties, action mechanism, and roles in dark repair. Mutat. Res. 236, 147-160.
Sancar, A. 1994 Structure and function of DNA photolyase. Biochemistry 33, 2-9.
Sancar, A. 1996 No "End of history" for photolyases. Science 272, 48-49.
Sancar, G.B. 2000 Enzymatic photoreactivation: 50 years and counting. Mutat. Res. 451, 25-37.
Sancar, A. 2008 Structure and function of photolyase and in vivo enzymology: 50th anniversary. J. Biol. Chem. 283, 32153-32157.
Sancar, G.B., F.W. Smith & P.F. Heelis 1987 Purification of the yeast PHR1 photolyase from an Escherichia coli overproducing strain and characterization of the intrinsic chromophores of the enzyme. J. Biol. Chem. 262, 15457-15464.
Sancar, G.B., F.W. Smith & A. Sancar 1985 Binding of Escherichia coli DNA photolyase to UV-irradiated DNA. Biochemistry 24, 1849-1855.
Schild, D., J. Johnson, C. Chang & R.K. Mortimer 1984 Cloning and mapping of Saccharomyces cerevisiae photoreactivation gene PHR1. Mol. Cell Biol. 4, 1864-1870.
Setlow, J.K. & R.B. Setlow 1963 Nature of the photoreactivable ultra-violet lesion in deoxyribonucleic acid. Nature 197, 560-562.
Setlow, J.K., M.E. Boling & F.J. Bollum 1965 The chemical nature of photoreactivable lesions in DNA. Proc. Natl. Acad. Sci. USA 53, 1430-1436.
Setlow, R.B. & W.L. Carrier 1964 The disappearance of thymine dimers from DNA: An error-correcting mechanism. Proc. Natl. Acad. Sci. USA 51, 226-231.
Setlow, R.B. & J.K. Setlow 1962 Evidence that ultraviolet-induced thymine dimers in DNA cause biological damage. Proc. Natl. Acad. Sci. USA 48, 1250-1257.
Setlow, R.B., W.L. Carrier & F.J. Bollum 1965 Pyrimidine dimers in UV-irradiated poly dI:dC. Proc. Natl. Acad. Sci. USA 53, 1111-1118.
Setlow, R.B., P.A. Swenson & W.L. Carrier 1963 Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142, 1464-1466.
Setlow, R.B., A.D. Woodhead & E. Grist 1989 Animal model for ultraviolet radiation-induced melanoma: Platyfish-swordtail hybrid. Proc. Natl. Acad. Sci. USA 86, 8922-8926.
Thiam, K. & A. Favre 1984 Role of the stringent response in the expression and mechanism of near-ultraviolet-induced growth delay. Eur. J. Biochem. 145, 137-142.
Thomas, G. & A. Favre 1975 4-Thiouridine as the target for near-ultraviolet light induced growth delay in Escherichia coli. Biochem. Biophys. Res. Commun. 66, 1454-1461.
Thomas, G., K. Thiam & A. Favre 1981 tRNA thiolated pyrimidines as targets for near-ultraviolet-induced synthesis of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem. 119, 381-387.
Todo, T., H. Takemori, H. Ryo, M. Ihara, T. Matsunaga, O. Nikaido, K. Sato & T. Nomura. 1993 A new photoreactivating enzyme that specifically repairs ultraviolet light-induced (6-4) photoproducts. Nature 361, 371-374.
Todo, T., H. Ryo, K. Yamamoto, H.Toh, T. Inui, H. Ayaki, T. Nomura & M. Ikenaga 1996 Similarity among the Drosophila (6-4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 272, 109-112.
Tsai, S-C., & J. Jagger 1981 The roles of the rel+ gene and of 4-thiouridine in killing and photoprotection of Escherichia coli by near-ultraviolet radiation. Photochem. Photobiology. 33, 825-834.
Van de Berg, B.J. & G.B. Sancar 1998 Evidence for dinucleotide flipping by DNA photolyase. J. Biol. Chem. 273, 20276-20284.
Varghese, A.J. & S.Y. Wang 1967 Ultraviolet irradiation of DNA in vitro and in vivo produces a third thymine-derived product. Science 156, 955-957.
Von Borstel, R.C. & S. Wolff 1955 Photoreactivation experiments on the nucleus and cytoplasm of the Habrobracon egg. Proc. Natl. Acad. Sci. USA 41, 1004-1009.
Weatherwax, R.S. 1956 Desensitization of Escherichia coli to ultraviolet light. J. Bacteriol. 72, 124-125.
Wulff, D.L. & C.S. Rupert 1962 Disappearance of thymine photodimer in ultraviolet irradiated DNA upon treatment with a photoreactivating enzyme from baker's yeast. Biochem. Biophys. Res. Commun. 7, 237-240.
Zelle, M.R. & A. Hollaender 1955 Effects of radiation on bacteria. Chap. 10 in A. Hollaender, Ed. Radiation Biology, Vol. II: Ultraviolet and Related Radiations (New York: McGraw-Hill), pp. 365-430.
11/8/08, Mod. 12/7/08

JOHN JAGGER
John Jagger died peacefully on December 27th. He was born in New Haven CT on 22 February 1924, the son of Carrie Eleanor Van Sickels of East Haven CT, and John William Jagger of New Haven CT. He had one brother, William Alexander Jagger, deceased 2000, of Hamden CT, and one sister, Ruth May Tolman, deceased 2004, of West Yarmouth MA. He studied physics at Yale College from 1946-1949, spent two years in the physics department of the Memorial-Sloan Kettering Cancer Center in New York City, and returned to Yale to get his PhD in Biophysics in 1954. He did post-doctoral research at the Radium Institute in Paris, and then worked nine years in the Biology Division of the Oak Ridge National Laboratory.
In 1965, he moved to Texas where he was Professor of Biology at the University of Texas at Dallas for 21 years. In 1956 in Oak Ridge he married Mary Esther Gaulden, radiation geneticist, who died in 2007. They had two children, Thomas Alexander Jagger of Austin TX, and Yvonne Callahan of Flower Mound, and three grandchildren, Alexander John Jagger, Melanie Nicole Mellinger and Kyle Allen Mellinger. John and Mary Esther lived with their family in Dallas from 1965 to Feb 2006, when they moved to Rambling Oaks Assisted Living in Highland Village TX.
He retired from UT-Dallas in 1986 to work and write on problems of science and society. In the 1990's, he served on the Board of directors of the Dallas Chapter of UNA-USA (United Nations), the Advisory Board of Advocates for Responsible Disposal in Texas (nuclear waste) and the Council of the North Texas Chapter, Health Physics Society. He was active in the Hillcrest Forest Neighborhood Association, helping to organize and lead a crime watch.
John was a biophysicist and photobiologist. He worked primarily on effects of ultraviolet light on bacteria. At UT-Dallas, working with a graduate student, T.V. Ramabhadran, they discovered the mechanism of growth delay induced by sunlight in bacteria. He was editor of the journal Photochemistry and Photobiology for three years, and President of the American Society for Photobiology (1983-84), which honored him in 1991 with a Lifetime Achievement Award. He wrote approximately 70 scientific papers and 4 scientific books, including The Nuclear Lion: What every Citizen should know about Nuclear Power and Nuclear War (plenum, 1991), and Science and the Religious Right: What Americans should know about Both (iUniverse, 2010).
He also published two personal Books: Cove Days: The Seaside Childhood of a Scientist (2002) and From Sea to Prairie: A Lifetime of Poems (2003). He loved aviation, trained and worked in aeronautical engineering during World War II, and in Texas become a private pilot, with instrument rating. He also liked aquatic sports, having grown up on the seashore, where he swam and sailed in his youth. He was especially proud of his back-yard swimming pool in Dallas, which he designed and swam in daily. The Family enjoyed many fourth of July parties there. With their children, they traveled to many places in the US, Europe and the Caribbean on 3 week vacation trips. They loved the outdoors and frequently hiked and camped. John enjoyed reading children's poetry and adventure books to his children and grandchildren. He and Mary Esther took yearly trips overseas from 1988-2002. They visited many countries in Europe, as well as the East Pacific, and went on an African safari. He was a member of the First Unitarian Church of Dallas and The American Humanist Association. There will be a memorial service on Sat. January 17th at 12:00 pm. at The First Unitarian Church of Dallas, 4015 Normandy Ave. Dallas, TX 75025. Donations may be made to The Nature Conservancy of Texas.
Published in Dallas Morning News on Jan. 8, 2015