PHOTOSYNTHETIC REACTION CENTERS
Charles Yocum
Departments of MCD Biology and Chemistry
University of Michigan, Ann Arbor, MI 49103-1048
cyocum@umich.edu
Translated into Macedonian
Introduction
Photosynthesis is initiated by a series of photochemical reactions in which light energy absorbed by chlorophylls is converted into redox energy that can be used to power a series of metabolic reactions. The reactions of light absorption in photosynthetic organisms can be divided into two separate processes. In the first, light is absorbed by antennae, proteins that bind several chlorophyll molecules. In many cases, a large number of antennae proteins are available to trap light energy. Chlorophylls that absorb light can pass on this energy to other adjacent pigments in a process called exciton transfer (i.e., resonance energy transfer). The structure and organization of antennae protein systems reveals that these proteins function as scaffolds that ligate their chlorophylls in highly organized complexes (Dekker and Boekema, 2005; Liu et al., 2004; McDermott et al., 1995). In many cases, the excitons generated in antennae are transferred to the second category of photoactive protein complexes that are known as reaction centers, the topic of this module.
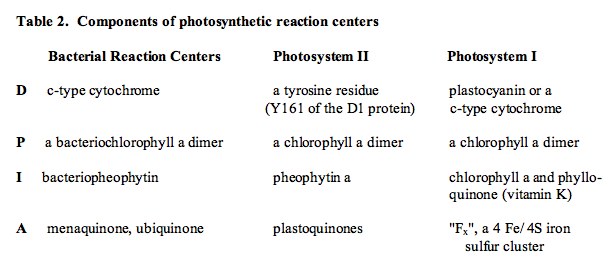
The photosynthetic reaction centers found in photosystems I and II of oxygenic organisms (cyanobacteria, red and green algae, higher plants), and in the photosynthetic bacteria that contain a single photosystem, are multisubunit membrane protein complexes that function as remarkable photochemical devices. They capture energy by either exciton transfer from antennae, or by direct absorption of light by chlorophyll. This light energy is then converted into oxidation-reduction potential energy, and is stabilized in the reaction center in a form that has a lifetime sufficiently long (milliseconds) to permit electrons to be extracted from the system. The transfer of these electrons to other redox enzymes is used to generate the ion and pH gradients that provide the energy for ATP synthesis, as well as the reducing power that is used for conversion of CO2 into sugars, starch and other metabolites. [See the module, Basic Photosynthesis, for a summary of energy conversion in photosynthetic membranes] All reaction centers have a remarkable similarity in their structure and composition. At their core, they consist of two intrinsic membrane proteins whose primary amino acid sequences are similar, but not identical. Interactions between these two proteins in all organisms form a heterodimeric structure that provides the binding sites for the cofactors that participate in the photochemical charge separation reactions. These cofactors include chlorophylls or bacteriochlorophylls, pheophytins and bacteriopheophytins (chlorophyll molecules lacking the central Mg2+ atom), and quinones (ubiquinone, plastoquinone or phylloquinone (vitamin K)).
The reactions that take place in a photosynthetic reaction center can be summarized by a model reaction sequence, in which the components are D (an electron donor molecule), P (the photocatalyst, a chlorophyll molecule), I (an intermediate electron carrier), and A (an electron acceptor molecule). The reaction can be written as follows:
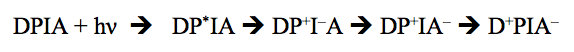
The state P* is the excited singlet state of the absorbing molecule, which in all cases is a chlorophyll. The succeeding reactions are very fast (times are given in Table 1). The species D+ and A-, which are the final products of the light-induced reaction, are highly reactive, differing in their reduction potentials by as much as 1.2 V. In spite of this enormous difference, they rarely react (recombine) with one another. This is because other oxidation-reduction reactions occur more rapidly and return the intermediate redox reactants to their original states, where they can participate in the next cycle of photochemical charge separation reactions. For example, in type II reaction centers (bacteria, photosystem II), an electron is transferred from A- to a second acceptor with a t1/2 of about 100-200 µsec, while in the same system D+ is reduced in a few µsec. The "secondary" recombination reactions, such as A- --> P+, are very slow compared to the rates of the forward electron transfer reactions.
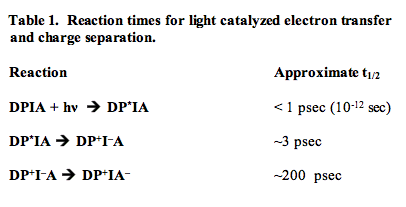
Table 2 lists the identities of D, P, I and A for several reaction centers. A common feature of these oxidoreductases is that the participants in the charge separation reactions are in all cases chlorophyll molecules, pheophytins, and quinone derivatives (a pheophytin is a chlorophyll molecule that does not contain the Mg2+ atom ligated in the center of the chlorin ring system). A biochemical analysis of the constituent proteins of reaction centers has shown that in all cases, the integral membrane proteins that ligate the electron transfer cofactors span the lipid bilayer.
The most important breakthrough in structural biology in the late 20th century was the crystallization of a bacterial reaction center, and the determination of its structure by Deisenhofer and Michel (Deisenhofer et al., 1994), who, along with Robert Huber, received the Nobel Prize in Chemistry for this achievement in 1988. Not only did this work open the door to the crystallization and solution of structures of a large number of diverse membrane proteins, but it also led finally to the crystallization of both photosystem I (PSI) and photosystem II (PSII), and the solution of their structures to resolutions of 2.5 and 1.9 Å, respectively. A number of interesting aspects of reaction center function have been revealed by these structures. A brief list is given below:
1. The reaction centers of bacteria, PSII and PSI possess a pseudo C2 symmetry; that is, one can divide centers in half to produce mirror images that are similar, but not identical;
2. As a consequence of the symmetry in reaction centers, they all possess two branches of electron transfer cofactors, either of which could, in theory, serve as an electron transport pathway;
3. The electron transfer cofactors are organized so that absorption of light translocates an electron across the lipid bilayer of the membrane.
Because the structure and function of the bacterial, or type II, reaction center is best characterized. The structure of this system will be used as the basis for understanding the function of all photosynthetic reaction centers.
The Reaction Center of Photosynthetic Bacteria
The structure of the reaction center from the photosynthetic bacterium Rhodopseudomonas viridis is shown in Figure 1. The complex is comprised of three proteins called "L", "M", and "H" (Low, Medium, High) after their apparent molecular masses on an SDS polyacrylamide gel. The L-M-H terminology is still in use, but DNA sequencing has resulted in a revision of the molecular masses, so that L-M-H is no longer accurate (L=31 kDa; M=34 kDa; H=28 kDa)). The "L" and "M" subunits have membrane spanning alpha-helices, and ligate the organic cofactors that make up the reaction center. A fourth subunit ligates the hemes that make up a cytochrome that functions as the electron donor to the oxidized reaction center bacteriochlorophylls.
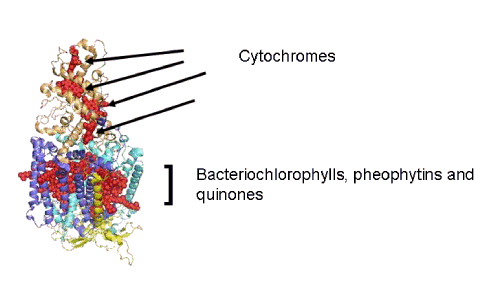
Figure 1. The reaction center from the photosynthetic bacterium Rhodopseudomonas viridis. The electron transfer cofactors are depicted as structures comprised of red spheres. The cytochromes supply electrons to a bacteriochlorophyll dimer called the "special pair" that absorbs light and reduces a bacteriopheophytin intermediate. There are four proteins, the "L" and "M" subunits (dark and light blue) whose alpha-helices span the membrane bilayer, the "H" subunit (yellow) on the side of the reaction center that accepts the electrons released by absorption of photons, and the cytochrome subunit that ligates the electron donor hemes (gold).
Figure 2 shows the cofactors of the reaction center without the proteins, and illustrates their mirror-image symmetry.
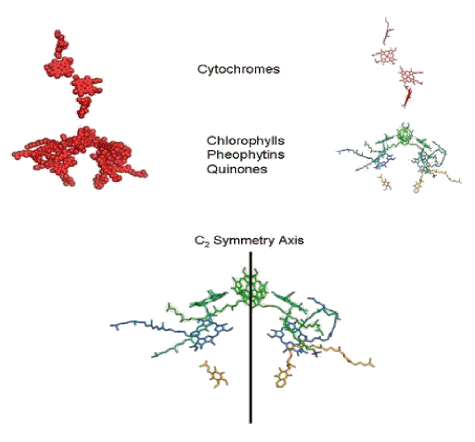
Figure 2. The cofactors of the bacterial reaction center, revealed by removal of the protein scaffolding. In the upper figure, the cofactors are shown as space-filling spheres (left), as they were depicted in Figure 1. On the right, one sees these cofactors represented as stick models. The hemes are red, bacteriochlorophylls are green, the bacteriopheophytins are blue, and the quinones are gold. In the lower figure, the cytochrome hemes have been removed, and a line has been drawn to show the approximate mirror-image symmetry of cofactors in the reaction center, which is called a C2 symmetry axis in the figure.
Figure 3 identifies the cofactors, and with this information, we are confronted with the big question. "Which set of cofactors transfers electrons after a light absorption event?" The answer is given away by the labeling used in Figure 3, where the arrows identify the molecules that comprise the "active" leg of the electron transfer chain in the reaction center.
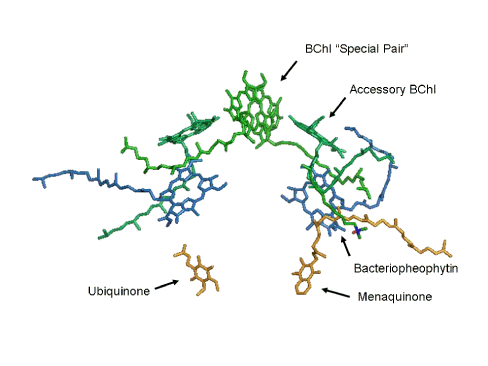
Figure 3. The organic cofactors of the reaction center. Four bacteriochlorophylls (BChl) are shown in green. One pair of these forms a dimer that is called the "special pair". Two bacteriopheophytins are shown in blue. Finally, the quinone electron acceptors, menaquinone and ubiquinone, and shown here in gold.
To illustrate how charge separation occurs along one "leg" of the reaction center, Figure 4 shows the events that are triggered when a photon is absorbed by the special pair of reaction center bacteriochlorophylls. The reacting species are identified sequentially by a red color in the structural model. The final, charge-separated state contains the oxidized special pair and a reduced ubiquinone. A second charge separation event and protonation yields a fully reduced ubiquinone that leaves its binding site in the reaction center. An oxidized quinone enters the vacant site so that charge separation can proceed. Reduced ubiquinone that is released from the reaction center is oxidized by a cytochrome bc1 complex in the bacterial membrane and the electrons return to the reaction center through the heme groups shown in Figures 1 and 2. Detailed diagrams of the cycle of reactions for bacterial photosynthesis can be found in recent biochemistry texts (Berg et al., 2001; Nelson and Cox, 2008; Voet and Voet, 2012).
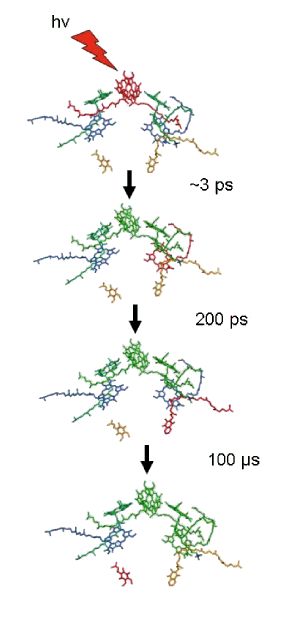
Figure 4. Light absorption triggers rapid charge separation in the bacterial reaction center. The pathway of electron transfer is indicated by the change in color to red as electron transfer proceeds through the cofactors.
The Photosystem II Reaction Center
The details about reaction center structure that were obtained from crystals of bacterial reaction centers provided the basis for theoretical models of the reaction center of photosystem II (PSII). Efforts to purify this reaction center showed that the cofactors were associated with a pair of proteins of similar amino acid sequences, called "D1" and "D2" on account of their diffuse appearance on SDS-polyacrylamide gels. A pair of small, membrane spanning polypeptides were also present that ligate the heme of a cytochrome, b559, whose function is still unclear. The number of chlorophyll a molecules in such a preparation was higher (six, rather than four as in bacteria), but two pheophytin a molecules were present, and a pair of plastoquinones were known to be present in this reaction center (Nelson and Yocum, 2006).
When the first crystals of PSII were obtained from thermophilic cyanobacteria (Thermosynechococcus elongatus and Thermosynechococcus vulcanus), it came as no surprise that the organization of the cofactors was shown to be very similar to those in the reaction centers of photosynthetic bacteria (Ferreira et al., 2004; Loll et al., 2005; Umena et al., 2011). There are outstanding differences as well. In the case of PSII, the electron donor is a tyrosine residue on the D1 protein, instead of the cytochromes that fulfill this function in bacteria. A continuous supply of electrons is available from H2O, which is oxidized by a cluster of inorganic ions (Mn, Ca, Cl). This topic is covered in the module on Oxygen Evolution. The PSII reaction center from T. elongatus is shown in Figure 5. Included among the cofactors shown are the hemes from cytochrome b559 and cytochrome c550, along with the inorganic ions that catalyze H2O oxidation and the redox-active tyrosine residue (called Yz) that shuttles electrons from Mn atoms to the oxidized special pair of chlorophyll a molecules, called P680 after the wavelength (in nanometers) where absorbance bleaching is observed after the absorption of a photon. The two peripheral chlorophyll a molecules (sometimes called chlorophyll D1 and chlorophyll D2) have been omitted from the figure to emphasize the similarities between PSII and the bacterial reaction center.
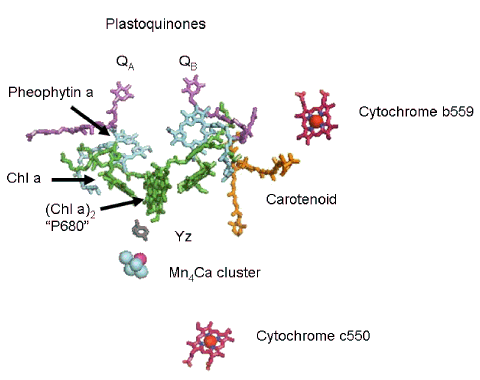
Figure 5. The photosystem II reaction center cofactors in T. elongatus. The special pair of chlorophyll a molecules (Chla)2 is also known as P680. A pair of chlorophyll a monomers is positioned between the special pair and the pheophytin a molecules shown in the structure. A pair of equivalent plastoquinone molecules, QA and QB, completes the inventory of organic cofactors. The pseudo C2 symmetry in this reaction center is disrupted by the presence of the inorganic cofactors shown here. Neither cytochrome is directly involved in primary electron transfer reactions. The Mn4Ca cluster catalyzes H2O oxidation, and along with the tyrosine residue YZ transfers electrons to P680 to reduce the special pair after it has absorbed a photon and transferred an electron to pheophytin a.
Figure 6 presents a diagram of the path of electron transfer in the PSII reaction center following absorption of a photon by P680. As in the case of the bacterial reaction center, one "leg" of the cofactor assembly is used to transfer the electron released from the special pair. The quinone binding site that accommodates QB can also be occupied by a group of herbicides, such as atrazine, which causes an inhibition of charge separation and is the basis of the phytotoxicity of such herbicides. Additional information on electron transfer in PSII can be found in the module on Oxygen Evolution.
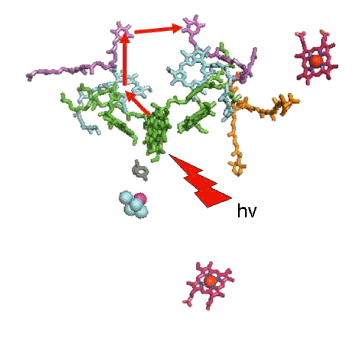
Figure 6. The pathway of electron transfer in the PSII reaction center. The series of reactions is identical to those in the bacterial reaction center, and is indicated by the red arrows. Double reduction and protonation releases QB (the quinone on the right) from its binding site, which then rebinds an oxidized plastoquinone molecule so that the next cycle of charge separation reactions can occur.
The Photosystem I Reaction Center
Biochemically-purified preparations of PSI reaction centers contain about 100 molecules of chlorophyll a. The reaction center chlorophyll in this photosystem, called P700 after the wavelength where absorption of a photon causes bleaching of absorbance, was proposed to be a dimer of chlorophylls based on the optical properties of synthetic chlorophyll dimers. When high resolution X-ray structures from PSI crystals from T. elongatus (Jordan et al., 2001) and from a higher plant (pea; Pisum sativa (Ben-Shem and Nelson, 2003)) became available, one was confronted with a large number of chlorophyll a molecules. Nevertheless, it was relatively easy to locate the position of the chlorophyll a dimer, other chlorophyll a species, the phylloquinones and the Fe/S clusters that make up the cofactors for charge separation in this reaction center. All of the cofactors are ligated to the same polypeptides that bind antenna chlorophyll a molecules and carotenoids. These proteins, called PsaA and PsaB, form the heterodimeric protein structure of the reaction center. The organization of cofactors in the PSI reaction center is shown in Figure 7.
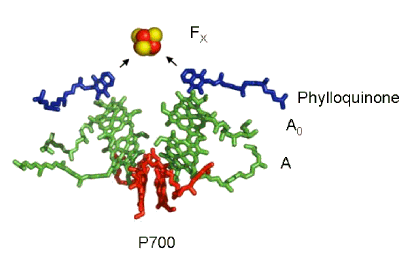
Figure 7. Cofactors of the PSI Reaction Center. The "special pair" of chlorophyll a molecules called P700 is shown in red. The other chlorophyll a species that are part of the pathway of electron transfer are shown in green, and the phylloquinone molecules are shown in dark blue. The primary electron acceptor is the four iron four sulfur cluster called FX, shown here as a red (iron) and yellow (sulfur) cube.
Once again, a symmetric disposition of the cofactors is revealed by the crystal structure, but this time there's a significant difference in the electron acceptor. In PSII and R. viridis a pair of quinone acceptors is present, and these function sequentially to accept electrons from the photochemical reaction catalyzed by the special pair of reaction center chlorophylls. In PSI, the cofactor "legs" come together at a single electron acceptor called FX, an iron-sulfur protein consisting of four Fe atoms and four inorganic sulfides. The present view is that excitation of P700 results in charge separation via electron transfer down either "leg" of cofactors (chlorophylls A and A0, phylloquinone). Figure 7 represents this by way of the two arrows, one from each "leg", directed at the primary electron acceptor FX. From FX, electrons are transferred to a pair of iron-sulfur clusters and are used to reduce the soluble iron-sulfur protein called ferredoxin, which donates the electrons to a flavoprotein that catalyzes reduction of NADP+ to NADPH. Photooxidized P700 is reduced by the water soluble copper protein called plastocyanin, or in some cases by a water soluble cytochrome. Electron transfer between PSII and PSI, which supplies the electrons needed to reduce P700+, is mediated by a complex containing cytochromes f and b6 and an iron-sulfur protein. Details of these reactions can be found in Berg, et al. (2001), Nelson and Cox (2008) and Voet and Voet (2012).
Summary
Photosynthetic reaction centers can be shown to have remarkably similar structures, comprised of two branches of cofactors that are made up of dimeric chlorophylls, in the case of light absorption, and either pheophytins or monomeric chlorophylls that are the components of the branches that transfer electrons to the ultimate electron acceptors, quinones or an iron-sulfur cluster. In the case of photosystem I, either branch can transfer electrons to the iron-sulfur cluster/acceptor. In bacteria and in photosystem II, only one branch is functional, and that branch delivers electrons to a tightly bound quinone, which then reduces a second, exchangeable quinone. The function of the "inactive" branch in the reaction centers is not obvious, and remains an interesting enigma concerning the structure and function of photosynthetic reaction centers.
Acknowledgement
Omri Drory and Nathan Nelson are thanked for their assistance with Figures 5-7.
References
Ben-Shem A, Frolow F, and Nelson N. The crystal structure of plant photosystem I. Nature 426, 630-635 (2003).
Berg, J.M., Tymoczko, J.L. and Stryer, L. Biochemistry. 5th ed. Chapter 19. W. H. Freeman and Company, New, NY (2001).
Dekker, J.P. and Boekema, E.J. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12-39 (2005).
Jordan, P., Fromme, P., Witt, H.T., Klukas, O., Saenger, W., Krauss, N.. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909-917 (2001).
Deisenhofer, J. and Michel, H., Epp, O., Sinning, I. and Michel, H. Crystallographic refinement at 2.3 Å resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis. J. Mol. Biol. 246, 429-457 (1995).
Ferreira, K.N., Iverson, T.M., Maghlaoui, K., Barber, J., and Iwata, S. Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831-1838 (2004).
Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., An, X., Chang, W. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287-292 (2004).
Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040-1044 (2005).
McDermott, G., Prince, S.M., Freer, A.A., Horthornthwaite-Lawless, A.M., Papiz, M.Z., Cogdell, R.J. and Isaacs, N.W. Crystal structure of an intergral membrane light-harvesting complex from photosynthetic bacteria. Nature 374, 517-521 (1995).
Nelson, D.L. and Cox, M.M. Principles of Biochemistry. 5th ed. Chapter 19. W. H. Freeman and Company, New, NY (2008).
Nelson, N. and Yocum, C.F. The structure and function of photosystems I and II. Ann. Rev. Plant Biol. 57, 521-565 (2006).
Umena, Y., Kwawkami, K., Shen, J.-R. and Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55-60 (2011).
Voet, D. and Voet, J. Biochemistry. 4th Ed., Chapt. 24, John Wiley and Sons, New York, NY (2012).
09/05/08
08/14/14