DNA DOUBLE-STRAND BREAKS
Kendric C. Smith
Emeritus Professor, Radiation Oncology (Radiation Biology)
Stanford University School of Medicine
800 Blossom Hill Road, Unit R169, Los Gatos, CA 95032
kendric@stanford.edu
web.stanford.edu/~kendric/
Introduction
DNA double-strand breaks (DSB) and single-strand breaks are NOT formed as a consequence of the direct absorption of UV radiation by DNA. Rather, they are formed as the consequence of the attempted repair of UV radiation-induced base damage in DNA.
Single-strand breaks are produced enzymatically during the incision step of the nucleotide excision repair of pyrimidine dimers, but these breaks are easily repaired by the action of DNA polymerase I and DNA ligase.
DSB are formed as the consequence of the production of excision repair breaks opposite each other on the two strands of DNA (Figure 1), and by the production of an excision repair break opposite a DNA daughter-strand gap (see module on Recombinational DNA Repair).

Figure 1. Model for the production of DNA DSB by overlapping nucleotide excision repair events. [Modified from Bonura and Smith, 1975b]
Formation of DNA Double-Strand Breaks
DSB appear in wild-type and polA1 strains of Escherichia coli after UV irradiation, and incubation in minimal growth medium. The uvrA6 strain, which is deficient in the incision step of nucleotide excision repair, showed no evidence of DSB under the same conditions. These results indicate that uvr+ cells, which are proficient in the incision step of nucleotide excision repair, accumulate DSB as a result of the excision repair process. Furthermore, the polA strain, which is deficient in DNA polymerase I, and is therefore deficient in excision repair subsequent to the incision step, showed more DSB than did the wild-type strain (Bonura and Smith, 1975a).
Normal human cells also show the formation of DSB after UV irradiation, but human cells deficient in excision repair (i.e., Xeroderma pigmentosum) do not show the formation of DSB (Bradley, 1981).
Unrepaired DNA DSB appear to be the major cause of lethality in UV-irradiated wild-type E. coli (Bonura and Smith, 1975a, b). Reducing the number of lesions in DNA after UV irradiation by the photoreactivation of pyrimidine dimers (see module on Photoreactivation), reduced the number of DSB observed (Bonura and Smith, 1975b).
Fluence fractionation also results in an increase in survival and a decrease in the yield of DSB (Bonura and Smith, 1975b). This result is consistent with the concept (Harm, 1968) that dose fractionation increases survival by decreasing the probability for the production of opposed excision gaps by distributing the production of the UV radiation damage over a longer time period during which DNA repair is operating.
Furthermore, for wild-type E. coli, the region of the UV radiation survival curve at which killing approaches exponential, corresponds to the fluence at which DSB accumulate linearly (Figure 2).
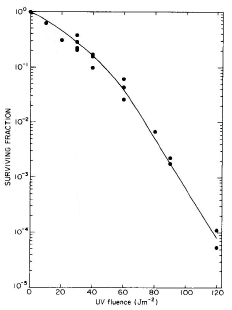

Figure 2. (A) Survival curve for log-phase E. coli K-12 cells grown and plated on minimal medium. (B) The appearance of DNA DSB as a function of UV fluence (254 nm). The straight line drawn through the 50-140 Jm-2 data is a least squares fit. [Modified from Bonura and Smith, 1975b]
Therefore, the type of DNA damage that ultimately results in cell death may be quite different from the initial radiation-induced lesion. In the present case, excisable pyrimidine dimers were converted to DNA single-strand breaks by repair enzymes, and subsequently to lethal DSB by the overlap of two DNA single-strand breaks, or by the overlap of DNA single-strand breaks with DNA daughter-strand gaps.
The repair of metabolically-produced DSB after UV irradiation constitutes a second type of recombination repair that is distinct from the repair of DNA daughter-strand gaps, i.e., the repair of DSB is RecBC-dependent, but the repair of DNA daughter-strand gaps is RecF-dependent (Wang and Smith,1983). (see module on Recombinational DNA Repair).
Repair of DNA Double-Strand Breaks
X-rays produce both DNA single-strand breaks and DSB as the consequence of the direct action of radiation on DNA. Therefore, most of what we know about the repair of DSB comes from work using ionizing radiation.
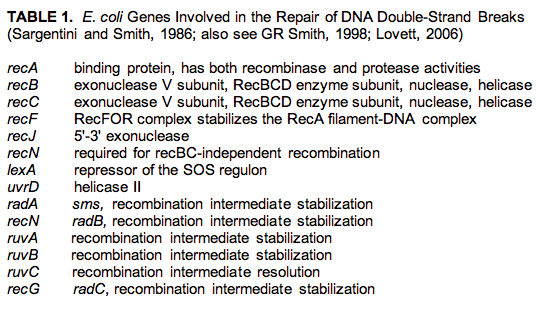
The precise steps in the repair of DSB are not well known, but a number of genes in E. coli are known to be involved in this type of repair
(Table 1).
Much of our knowledge about the repair of DSB comes from studies on X-ray-induced mutagenesis, especially the production of long deletion mutations.
Experiments designed to determine the association between the repair of gamma-radiation-induced DSB and the induction of 700-1000 bp long deletions (Lac- --> Lac+), base substitutions (leuBl9 --> Leu+), and frameshifts (trpE9777 --> Trp+) in E. coli K-12, showed that over the range of 2.5-20 krad, deletions were induced with linear kinetics, as has been observed for the induction of DSB, while the induction kinetics for base substitutions and frameshifts were curvilinear.
Both the repair of DSB, and the induction of deletion mutations showed an absolute requirement for an intact recB gene, as well as a dependency on the type of preirradiation growth medium used (i.e., rich medium vs. minimal medium). These requirements were not seen for base substitutions or frameshifts (Sargentini and Smith, 1992). (see module on UV Radiation and Spontaneous Mutagenesis)
This dependency on rich growth medium for enhanced survival is thought to result from the greater number of sister-DNA duplexes present, and the more rapid synthesis of RecA protein in cells grown in rich medium versus minimal medium (Sargentini et al., 1983).
It has been proposed that the RecBCD enzyme attaches to the end of the DSB, and unwinds (and rewinds) the DNA duplexes until a Chi site in encountered. Chi sites are a sequence of bases (5'-GCTGGTGG-3') positioned about every 5 Kb throughout the DNA, and are recombination hot spots. Chi is a recognition site for the RecBCD enzyme (Exonuclease V), which nicks DNA near Chi as it unwinds DNA (GR Smith and Stahl, 2005).
Since the unwinding action is more rapid than the rewinding action, single-stranded DNA loops are formed. Cutting of the single-stranded DNA loop near the Chi site allows the invasion of the 3' end into a sister-DNA duplex, where it can search for a homologous DNA sequence. The proper alignment of the invading strand with its complementary homologue can lead to the accurate repair of DSB. However, misalignment of the invading strand at another locus would lead to a loop of non-homologous single-stranded DNA that could be lost by excision, or through bypass replication, thus producing a deletion (Sargentini and Smith, 1992).
The repair of DSB in E. coli requires recombinational repair systems, i.e., two DNA duplexes are required, and strand replacement occurs by homologous recombination (see Friedberg et al., 2006; p. 579). However, in mammalian cells, some DSB are repaired by non-homologous end joining (Ferguson et al., 2000).
Summary and Conclusions
This module concentrated on the fact that UV radiation-induced DNA DSB are not formed as the consequence of the direct absorption of UV radiation by DNA, but are formed AFTER UV irradiation by the attempted nucleotide excision repair of UV radiation-induced pyrimidine dimers. It should be noted that DNA DSB are also formed during the repair of other types of DNA damage, e.g., mismatch repair (Nowosielska and Marinus, 2008), and replication arrest (Michel et al., 1997).
Video: Homologous Recombination of DNA Double-Strand Breaks
References
Bonura T, Smith KC. 1975a. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J Bacteriol 121:511-517.
Bonura T, Smith KC. 1975b. Quantitative evidence for enzymatically-induced DNA double-strand breaks as lethal lesions in UV-irradiated pol+ and polA1 strains of E. coli K-12. Photochem Photobiol 22:243-248.
Bradley, MO. 1981. Double-strand breaks in DNA caused by repair of damage due to ulatraviolet light. J Supramolec Struct Cell Biochem 16:337-343.
Ferguson, DO, Sekiguchi, JM, Chang, S, Frank, KM, Gao, Y, DePinho, RA, Alt, FW. 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. PNAS 97: 6630-6633.
Friedberg, EC, Walker, GC, Siede, W, Wood, RD, Schultz, RA, Ellenberger, T. 2006. Recombinational Repair, Replication Fork Repair, and DNA Damage Tolerance, Chapter 16, pp. 569-612, DNA Repair and Mutagenesis (2nd ed.), ASM Press, Washington, D.C.
Harm, W. 1968. Effects of dose fractionation on ultraviolet survival of Escherichia coli. Photochem. Photobiol. 7:73-86.
Lovett, ST. 2006. Replication arrest-stimulated recombination: Dependence on the RecA paralog, RadA/Sms and translesion polymerase, DinB. DNA Repair (Amst). 5:1421-1427.
Michel, B, Ehrlich, SD, Uzest, M. 1997. DNA double-strand breaks caused by replication arrest. The EMBO Journal 16:430-438.
Nowosielska, A, Marinus, MG. 2008. DNA mismatch repair-induced double-strand breaks. DNA Repair 7:48-56.
Sargentini, NJ, Diver, WP, Smith KC. 1983. The effect of growth conditions on inducible recA-dependent resistance to X rays in Escherichia coli. Radiat. Res. 93:364-380.
Sargentini, NJ, Smith KC. 1986. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat. Res. 107:58-72.
Sargentini, NJ, Smith, KC. 1992. Involvement of RecB-mediated (but not RecF-mediated) repair of DNA double-strand breaks in the gamma-radiation production of long deletions in Escherichia coli. Mutat. Res. 265:83-101.
Smith, GR, 1998. DNA double-strand break repair and recombination in Escherichia coli. In DNA Damage and Repair: DNA Repair in Prokaryotes and Lower Eukaryotes. Nickoloff, JA, Hoekstra, MF, (eds), Humana Press, pp. 135-162.
Smith, GR, Stahl, FW. 2005. Homologous recombination promoted by Chi sites and RecBC enzyme of Escherichia coli. Bioessays 2:244-249.
Wang, TV, Smith KC. 1983. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol 156:1093-1098.
AND SCROLL TO END OF MODULE.
[NOTE: Papers by the author are available as PDF files.]
10/09/08
06/02/11
03/19/14