PHOTOMOVEMENT of BACTERIA and ARCHAEA
Wouter D. Hoff
Department of Microbiology and Molecular Genetics
Oklahoma State University, Stillwater OK 74078
wouter.hoff@okstate.edu
1. Overview
Studies on the phototactic and chemotactic responses in a number of bacteria and archaea have yielded two basic mechanisms of action for prokaryotic phototaxis: (i) responses triggered via the bioenergetic consequences of photoexcitation of the photosynthetic machinery, and (ii) responses triggered by dedicated photosensory receptors (Figure 1).
Responses of the first type may involve changes in redox potential, caused by photosynthetic electron transport, or changes in proton motive force. These changes in redox potential or membrane potential are then sensed by, as yet unidentified, sensors, and relayed to the locomotive machinery, often the bacterial flagellum. This mode of phototaxis is exemplified by the phototaxis responses of Rhodobacter sphaeroides (Berry and Armitage 2000).
In the second case, photoexcitation of a dedicated photoreceptor results in the conversion of the initial state of the photoreceptor to its signaling state. The molecular mechanism of photoreceptor activation in both the archaeal sensory rhodopsins (Hoff 1997), and the eubacterial blue light receptor photoactive yellow protein (Meyer 1987, Sprenger 1993) have attracted extensive studies as model systems for understanding receptor activation and functional protein dynamics in general. Formation of the signaling state then relays a signal into an associated signal transduction chain, leading to the bacterial flagellum (Figure 1). The model system for this type of phototaxis is formed by the sensory rhodopsins in the halophilic archaeon Halobacterium salinarum (Hoff 1997).
Recently, two other classes of photoreceptors have been identified as photoreceptors for prokaryotic photomovement, in addition to the archaeal sensory rhodopsins and photoactive yellow protein: the bacterial phytochromes (Davis 1999) and BLUF flavoproteins (Gomelski 2002), with AppA as the founding member (Gomelsky 1998; Masuda and Bauer 2002). The phototaxis responses in Synechocystis sp. PCC6803, which are based on twitching motility, involve both the BLUF protein PixD (Okajima 2005) and the phytochrome-like proteins pixJ1 (Yoshihara 2000; Bhaya 2001) and Cph2 (Wilde 2002). The LOV flavoproteins (Briggs 2001, 2007; Christie 1999; Crosson 2003) and cryptochromes (Sancar 2003) have not yet been reported as the photoreceptors for a prokaryotic phototaxis response. Thus, currently four different types of photosensory proteins are involved in prokaryotic phototaxis: the sensory rhodopsins in halophilic archaea, PYP in Halorhodospira halophila, and the BLUF protein PixD and phytochrome-related proteins PixJ1 and Cph2 in Synechocystis.
In the final stage of signaling in prokaryotic phototaxis, the signal transduction chain interacting with the dedicated photoreceptor or sensor of bioenergetic status affects the locomotive machinery of the bacterium. This often involves changes in the frequency with which the bacterial flagellum changes its direction of rotation. However, other phototactic bacteria utilize gliding motility (McBride 2001), or movement of entire colonies of bacteria (Ragatz 1994, 1995). These photomovement are based on changes in swarming (Ragatz 1995) or twitching (Ng 2003) motility based on type IV pili.
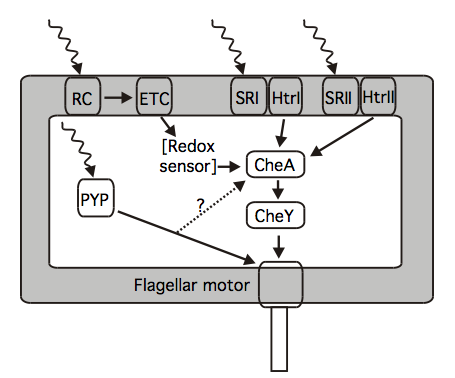
Figure 1. Summary of available knowledge on the components in the phototaxis signaling pathways of Halobacterium salinarum, Rhodobacter sphaeroides, and Halorhodospira halophila. H. salinarum contains the photoreceptors SRI and SRII, which are complexed in the membrane to their signal transducers HtrI and HtrII. These transducers modulate the autokinase activity of CheA, and thus modulate the phosphorylation status of CheY. Phototaxis of R. sphaeroides proceeds via its photosynthetic reaction center (RC) and electron transfer chain (ETC) via a putative redox sensor. Positive phototaxis in H. halophila occurs via a similar pathway, while its negative phototaxis is triggered by photoactive yellow protein (PYP). The signal transduction pathway for PYP is unknown; one candidate is the Che system. Possible adaptation mechanisms are not shown. [Adapted from (Hoff 2009)]
The photoreceptors triggering photomovement in prokaryotes will first be discussed, followed by the signal transduction chain associated with these photoreceptors, and finally a summary of the changes in prokaryotic motility triggered by light stimuli. A number of reviews on these topics are available (Hader 1987; Armitage 1997; Sgarbasa 2002; Armitage and Hellingwerf 2003; Van der Horst and Hellingwerf 2004; Hoff 2009).
2. Photoreceptors
2a. Phototaxis via the photosynthetic machinery.
i. Phototaxis via photosynthetic electron transport: redox sensing. Many free-swimming purple photosynthetic bacteria, such as Rhodobacter sphaeroides, Rhodocista centenaria (formerly Rhodospirillum centenum), and Halorhodospira halophila (formerly Ectothiorhodospira halophila), accumulate in areas illuminated with photosynthetically active light (Hustede 1989; Sprenger 1993; Sackett 1997; Ragatz 1995; Gest 1995). The photoreceptor for this positive phototactic response is the photosynthetic machinery. Illumination initiates photosynthetic electron transport, and a presumed redox sensor then relays the signal to the flagellar motor (Armitage 1997; Grishanin 1997). Thus, at the heart of this type of phototaxis is a redox sensing mechanism, and the redox sensor is coupled to the photosynthetic machinery by the photosynthetic electron transfer chain. The proteins involved in this redox sensing mechanism have not yet been identified.
ii. Phototaxis via bacteriorhodopsin: proton motive force sensing. The halophilic archaebacterum Halobacterium salinarum exhibits various phototactic responses. These are triggered by the two dedicated photoreceptors, sensory rhodopsin I and II (see below). However, mutants of H. salinarum lacking these two photoreceptors still exhibit residual phototaxis responses. These are transmitted by the transmembrane proton pump bacteriorhodopsin (Bibikov 1993). Evidence has been reported to show that these bacteriorhodopsin-based phototaxis responses are mediated by a "protometer" that senses the transmembrane electrochemical gradient (Grishanin 1996). The protein(s) constituting the protometer have not yet been identified.
2b. Dedicated photoreceptors.
Most knowledge on the structure, dynamics, and function of dedicated prokaryotic photoreceptors has been derived from studies of two systems: the sensory rhodopsins (SRs) in the archaeon Halobacterium salinarum (Hoff 1997), and photoactive yellow protein (PYP) from the eubacterium Halorhodopsira halophila (Cusanovich and Meyer 2003; Helingwerf 2003; Imamoto and Kataoka 2007). In a striking case of convergent evolution, these two extremely halophilic organisms have evolved completely different photosensory systems that exhibit remarkably close analogies in their molecular functioning. In addition, recently a rapidly growing body of knowledge on bacterial phytochromes (Davis 1999; Wagner 2005) and BLUF proteins (Gomelski 1998, 2002; Masuda and Bauer 2002; Anderson 2005; Jung 2005; Kita 2005) has been reported.
i. Archaeal sensory rhodopsins. Halobacterium salinarum is an extremely halophilic organism that can grow both aerobically and, under conditions of illumination and reduced oxygen tension, photosynthetically. H. salinarum exhibits both positive and negative phototactic responses (Spudich and Bogomolni 1988; Marwan and Oesterhelt 1990). The photosynthetic and phototactic capabilities of this archaeon are based on four different rhodopsins. First, H. salinarum contains two light-driven ion pumps: bacteriorhodopsin (BR, a transmembrane proton pump) and halorhodopsin (HR, a transmembrane chloride pump). Secondly, it contains two separate archaeal rhodopsins (SRI and SRII) that cause distinct phototactic responses (Hoff 1997), as discussed below. SRII is also referred to as phoborhodopsin. The alkalophilic archaeon Natronobacterium pharaonis contains a photoreceptor very similar to the SRII from H. salinarum (Engelhard 1996). The Natronobacterium SRII has proven to be amenable X-ray crystallographic studies (see below). Results from on the sensory rhodopsins from both archaea have led to a unified view of these photosensors (Hoff 1997; Sasaki and Spudich 2008).
All four Halobacterial rhodopsins (Table 1) consist of a bundle of seven transmembrane (
 -helices (labeled A to G), and contain a covalently linked retinal chromophore embedded in these helices (Figure 2). They share ~25% amino acid sequence identity, particularly in the retinal binding pocket, formed by the residues that are immediately adjacent to the retinal molecule. Thus, initial models for the structure of the sensory rhodopsins were based on the structure of BR as derived from cryoelectron microscopy experiments using the naturally occurring two-dimensional crystals of BR (Grigorieff 1996). Recently, a wealth of structural data has been obtained on these proteins: X-ray structures of BR at 1.55 Å resolution (Luecke 1999), HR at 1.8 Å resolution (Kolbe 2000), SRII at 2.1 Å resolution (Luecke 2001; Royant 2001) and SRII in complex with the transmembrane part of its signal transducer HrtII at 1.94 Å (Gordeliy 2002). These results confirmed the high degree in similarity of the four archaeal rhodopsins and their retinal binding pockets, while also revealing subtle but functionally crucial differences in their structures. A detailed analysis of the structure-function relationships in these proteins in now ongoing, and promises to bring our understanding of the physical principles that are involved in the function of these proteins to a new level. One key recent advance is the demonstration that only three point mutations in bacteriorhodopsin result in a protein with sensory properties resembling those of the sensory rhodopsins (Sudo and Spudich 2006).
-helices (labeled A to G), and contain a covalently linked retinal chromophore embedded in these helices (Figure 2). They share ~25% amino acid sequence identity, particularly in the retinal binding pocket, formed by the residues that are immediately adjacent to the retinal molecule. Thus, initial models for the structure of the sensory rhodopsins were based on the structure of BR as derived from cryoelectron microscopy experiments using the naturally occurring two-dimensional crystals of BR (Grigorieff 1996). Recently, a wealth of structural data has been obtained on these proteins: X-ray structures of BR at 1.55 Å resolution (Luecke 1999), HR at 1.8 Å resolution (Kolbe 2000), SRII at 2.1 Å resolution (Luecke 2001; Royant 2001) and SRII in complex with the transmembrane part of its signal transducer HrtII at 1.94 Å (Gordeliy 2002). These results confirmed the high degree in similarity of the four archaeal rhodopsins and their retinal binding pockets, while also revealing subtle but functionally crucial differences in their structures. A detailed analysis of the structure-function relationships in these proteins in now ongoing, and promises to bring our understanding of the physical principles that are involved in the function of these proteins to a new level. One key recent advance is the demonstration that only three point mutations in bacteriorhodopsin result in a protein with sensory properties resembling those of the sensory rhodopsins (Sudo and Spudich 2006).

The structure of these four aechaeal rhodopsins clearly resembles that of the large and important eukaryotic family of G-protein coupled receptors (GPCRs), which share the seven-transmembrane-helix structural motif. The visual rhodopsins in the eyes of animals are a part of this family, but so are a range of other receptors. The human genome is estimated to contain over 1,000 GPCRs. Over 50% of all modern drugs target GPCRs (Guderman 1995), and 25% of the most utilized dugs bind to this class of receptors (Flower 1999). The archaeal rhodopsins serve as biophysically accessible model systems for this important class of proteins.
Photoreceptor proteins derive their ability to absorb light above 300 nm from bound light-absorbing cofactors, or chromophores. The functionally active conformation of the chromophore in the dark state of the protein of all four archaeal rhodopsins is all-trans retinal, covalently attached to a highly conserved Lys side chain via a protonated Schiff base linkage (Figure 2). The protonated Schiff base is fully enclosed by the protein matrix, and thus represents a buried charge. This charge is stabilized by electrostatic interactions with nearby charged residues. This has been investigated most extensively for BR, where the counterion for the protonated Schiff base is thought to consist of two negatively charged side chains (Asp85 and Asp96) and one positively charged side chain (Arg82) that interact with the Schiff base via a water molecule (Marti 1991; Maeda 1994). A similar situation, but with significant modifications, is found in the other archaeal rhodopsins.
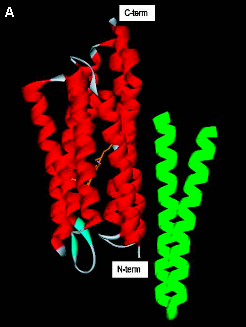
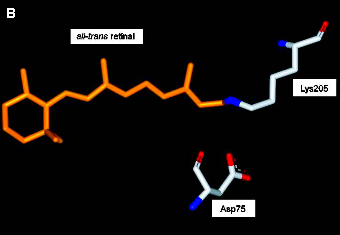
Figure 2. Structure of sensory rhodopsin II.
(A) The overall structure of SRII (transmembrane-helices in red) complexed to the transmembrane
-helices of HtrII (green).
(B) The negatively charged side chain of Asp76 forms a salt bridge with the protonated Schiff Base linking the retinal chromophore to Lys205. (Based on Gordeliy 2002).
In spite of the fact that the above four archaeal rhodopsins share an identical chromophore, their absorbance spectra vary significantly (see Table 1). The phenomenon that a protein binding pocket alters the absorbance spectrum of the chromophore is referred to as spectral tuning. Particularly because spectral tuning in the visual rhodopsins in our eyes is the basis of our color vision, this effect has received significant attention. Previous experimental studies on the visual rhodopsins and bacteriorhodopsin, and computational work based on the high-resolution structures of BR and SRII have yielded a view in which the following effects are the main contributors to spectral tuning in the rhodopsins (Ren 2001; Hayashi 2001; Yan 1995; Hu 1994; Kochendoerfer 1999). (i) The effective distance between the protonated Schiff base and its counterion, where a shorter distance results in a more blue-shifted absorbance spectrum. (ii) The conformation of the retinal, particularly twists around single bonds. Especially the C6-C7 single bond plays an important role in this regard. In the case that the retinal chain and its
 -ionone ring are co-planar, a more red-shifted absorbance spectrum results, because of the effective lengthening of the conjugated system. (iii) Stabilization of the electronic excited state by polar or polarizable side chains located along the retinal polyene chain (Yan 1995; Kochendoerfer 1999). However, important questions on the detailed physical basis of spectral tuning in the rhodopsins remain, e.g., compare (Ren 2001 and Hayashi 2001).
-ionone ring are co-planar, a more red-shifted absorbance spectrum results, because of the effective lengthening of the conjugated system. (iii) Stabilization of the electronic excited state by polar or polarizable side chains located along the retinal polyene chain (Yan 1995; Kochendoerfer 1999). However, important questions on the detailed physical basis of spectral tuning in the rhodopsins remain, e.g., compare (Ren 2001 and Hayashi 2001).
Photoexcitation of all four archaeal rhodopsins results in the ultrafast all-trans to 13-cis photoisomerization of the retinal chromophore on a time scale of 0.5 to 5 picoseconds (Kennis and Groot 2007). Thus, double bond photoisomerization is the primary chemical event that initiates the function of these proteins. This initial process involves relatively small changes in the structure of the retinal, in which the strain of fitting the 13-cis chromophore in the all-trans binding pocket is distributed over a number of bond angles. A significant fraction of the energy represented by the absorbed photon is stored in the initial photoproduct (~80 kJ/mol) (Birge and Cooper 1983). This provides the driving force for a series of thermal reactions that finally result in the reformation of the initial state. This cyclic chain of thermal transitions is referred to as a photocycle. The photocycle of BR and HR is completed in ~10 milliseconds, and results in the transmembrane translocation of a proton and chloride ion, respectively. In the SRs the photocycle is significantly slower: recovery of the initial state of the receptor requires ~1 second. During this photocycle the protein is converted from its initial receptor state to a signaling state that is active in relaying a signal into the associated downstream signal transduction chain.
The photocycle of the archaeal SRs has been studied in some detail (Spudich and Bogomolni 1988; Hoff 1997), and is schematically depicted in Figure 3. While these photocycles generally resemble that of the well-studied BR photocycle, some important differences exist. First, the time scale of the photocycles of the SRs is ~100-fold slower than that of BR. This difference matches the sensory function of the SRs: the lifetimes signaling state(s) generated during their photocycle need to be sufficient for efficient relay of a signal into the attached signal transduction chain. Secondly, in the photocycle of SRI, red-shifted N- and O-like intermediates are not observed in the last part of the photocycle. The main three stages of the photocycles of the SRs are (i) retinal isomerization to yield an initial photoproduct, (ii) deprotonation of the Schiff base, resulting in a blue-shifted intermediate, and (iii) recovery of the initial state of the receptor. The Schiff base deprotonation step is readily identified by time-resolved UV/vis absorbance spectroscopy, since it results in a strong blue-shift of the absorbance maximum of the protein to around 370 nm.
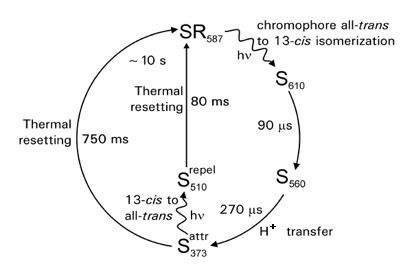
Figure 3. Schematic representation of the photocycle of sensory rhodopsin I. Light-triggered transitions are indicated by wavy arrows, thermal transitions by straight arrows. Absorbance maxima are shown as subscripts. The time constants and main molecular processes for the transitions are indicated. The formation of S373 elicits an attractant response, and the formation of S510 a repellent response.
The proton transfer pathway involved in the deprotonation and reprotonation of the retinal Schiff base during the photocycle has received significant attention. For SRI in the absence of its transducer (see below) Asp76 functions as the acceptor for the Schiff base proton (Rath 1994, 1996). This strongly resembles the role of Asp85 during the proton pumping photocycle of bacteriorhodopsin. However, in the presence of the transducer for SRI, which is the physiological situation, Asp76 is protonated, and an unidentified residue accepts the Schiff base proton during the SRI photocycle. Unexpectedly, His166 plays an important role in these proton transfer reactions of SRI (Zhang and Spudich 1997). This is in contrast to the proton transfer pathway in BR, where no His side chains are involved. For SRII it has been found that Aps73 is the acceptor for the Schiff base proton during the photocycle. The proton transfer from the Schiff base to Asp73 disrupts the active site salt bridge between these two residues, an event that is though to be central to the receptor activation process in SRII. An important observation was made for the D73N mutant of SRII. In this mutant, the salt bridge between the protonated Schiff base and residue 76 is already disrupted before photoexcitation of the photoreceptor. The D73N mutation results in the constitutive activation of SRII (Spudich 1997). This mutation is highly reminiscent of the E113Q mutation in human rhodopsin, which also results in constitutive signaling. Thus, the light-triggered disruption of an active site salt bridge is a general mechanism for the activation of rhodopsin photoreceptors. A key element in signaling by sensory rhodopsin I is a conformational change that determines the connectivity of the Schiff base to either the cytoplasm or the periplasm (Sineshchekov 2008). Interestingly, this connectivity switch determines if SRI photoexcitation causes an attractant or a repellent response.
Excitation of SRI results in a positive phototaxis response. The functional relevance of this response is thought to be accumulation of the cells in light that excites the proton pump BR. This role is in line with the expression of SRI under conditions where BR is also expressed. A functionally central feature of the photocycle of SRI is that the blue-shifted S373 intermediate exhibits efficient photochemistry upon absorption of near-UV light. This results in the formation of a novel intermediate, which triggers a negative phototaxis response. Thus, the single protein SRI is responsible both for positive phototaxis towards light around 590 nm, and for negative phototaxis towards light around 370 nm. This constitutes a mechanism of color discrimination (Spudich and Bogomolni 1984). Note that this negative phototaxis response only occurs in the presence of 590 nm light, which results in the formation of the S373 state.
SRII is expressed under conditions where BR is not present in the cell. Its photoexcitation triggers a negative phototaxis response away from 490 nm light, and is thought to keep cells away from potentially damaging light when the light-driven ion pumps BR and SR are not synthesized, and the cell thrives by aerobic, non-photosynthetic growth.
The main functional feature of the photocycles of SRI and SRII is to convert the initial state of the receptor, which does not relay a signal into the downstream signaling chain, into a conformation of the receptor, its signaling state, that is active in downstream signal relay. A set of important and unique experiments has been performed on the Halobacterial SRs, which allowed the in vivo identification of the signaling states of SRI and SRII (Yan and Spudich 1991; Yan 1991). This experiment involved the in vivo reconstitution of retinal-deficient cells with various retinal analogues. These retinal analogues differently affect the various time constants in the SR photocycles. By relating the effects that these analogues have on the photocycle kinetics with changes in the phototactic sensitivity of the cells towards light, the functional signaling states could be identified as S373 in SRII and S350 and S530 in SRI. These experiments provide direct experimental verification for the validity of the concept of the signaling state in the living cell.
ii. Photoactive yellow protein. The purple sulfur bacterium Halorhodopsira halophila is an obligately photosynthetic and extremely halophilic eubacterium that contains photoactive yellow protein (PYP). For reviews on the properties of PYP see (Cusanovich and Meyer 2003; Hellingwerf 2003; Imamoto and Kataoka 2007). The PYP from H. halophila causes a negative phototaxis response of this bacterium to blue light based on the observation that the wavelength dependence of this response matches the absorbance spectrum of PYP (Sprenger 1993). However, because the role of PYP as the photoreceptor has not yet been confirmed by genetic tools, its function remains tentative.
H. halophila exhibits not only negative phototaxis to blue light, but also positive phototaxis towards photosynthetically active light (Hustede 1989; Sprenger 1993). While the negative phototaxis response is thought to occur via PYP, the positive phototaxis response likely involves a mechanism similar to that in Rhodobacter sphaeroides (see Section 2a). The positive phototaxis response attracts the cells into areas where photosynthesis is possible; the negative phototaxis response is believed to repel cells from areas with excessive, potentially damaging light intensities.
PYP is also found in a number of other purple photosynthetic bacteria, including Chromatium salexigens, Rhodopsirillum salexigens, Rhodocista centenaria, Rhodobacter sphaeroides, and Rhodobacter capsulatus, and in the Bacteriodetes Salinibacter ruber (Kort 1996b; Kumauchi 2008; Memmi 2008) These proteins form a well-conserved family of photoreceptor proteins. Note, however, that the PYP homolog from R. centenaria is part of a much larger protein containing phytochrome and histidine kinase domains, and is thought to regulate gene expression (Jiang 1999). Thus, there appears to be functional variability within the PYP photoreceptor family.
PYP is small (125 amino acids) and water soluble protein with a strong yellow color (
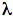 max = 446 nm,
max = 446 nm,  446 = 45.5 mM-1cm-1). The chromophore of PYP responsible for its yellow color is p-coumaric acid, covalently linked to a conserved Cys residue in the protein via a thioester bond (Figure 4)(Hoff 1994; 1996; Baca 1994; van Beeumen 1993). This represented the first time that p-coumaric acid was found as a protein cofactor, establishing PYP as a novel type of photoreceptor.
446 = 45.5 mM-1cm-1). The chromophore of PYP responsible for its yellow color is p-coumaric acid, covalently linked to a conserved Cys residue in the protein via a thioester bond (Figure 4)(Hoff 1994; 1996; Baca 1994; van Beeumen 1993). This represented the first time that p-coumaric acid was found as a protein cofactor, establishing PYP as a novel type of photoreceptor.
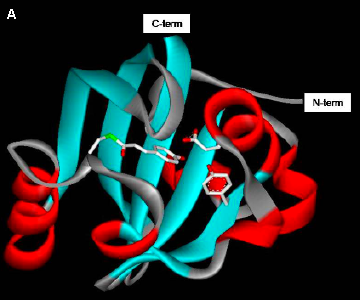
Figure 4. Structure of photoactive yellow protein.
(A) Cartoon representation of the structure of a PixD monomer, with-helices in red and
-sheets in blue.
(B) The p-coumaric acid chromophore is buried within the protein and its negatively charged phenolic oxygen is hydrogen bonded with the side chains of Tyr42 and Glu46. (Based on (Borgstahl 1995).)
The pyp gene from H. halophila has been successfully overexpressed in E. coli, followed by in vitro reconstitution of the holoprotein (Imamoto 1995; Kort 1996b). This has greatly facilitated biophysical studies on the protein. The structure of the PYP from H. halophila has been determined at high resolution both by X-ray crystallography (Borgstahl 1995; Getzoff 2003) and multidimensional NMR spectroscopy (Dux 1998). This revealed that PYP consists of a central 6-stranded
 -sheet, flanked on both sides by
-sheet, flanked on both sides by  -helices (Figure 4). Interestingly, this structure is common to a large number of domains that are all involved in signaling and regulation. This family of proteins is referred to as PAS domains (Taylor and Zhulin 1999). In addition, the crystal structure of PYP yielded detailed information on the amino acid side chains in the chromophore binding pocket. The crystal structure of the PYP homolog from R. centenaria was shown to be largely similar to the PYP from H. halophila (Rajagopal and Moffat 2003).
-helices (Figure 4). Interestingly, this structure is common to a large number of domains that are all involved in signaling and regulation. This family of proteins is referred to as PAS domains (Taylor and Zhulin 1999). In addition, the crystal structure of PYP yielded detailed information on the amino acid side chains in the chromophore binding pocket. The crystal structure of the PYP homolog from R. centenaria was shown to be largely similar to the PYP from H. halophila (Rajagopal and Moffat 2003).
Like the rhodopsins, PYP exhibits significant spectral tuning of the absorbance spectrum of its chromophore. Free p-coumaric acid at neutral pH has an absorbance maximum at 284 nm, while the absorbance maximum of native PYP is at 446 nm. This strong red-shift has been dissected into chemical effects and physical effects caused interactions between the chromophore and its binding pocket. The chemical factors (Kroon 1996) consist of the formation of the thioester bond (shift to 335 nm), and the deprotonation of the phenolic oxygen of the chromophore (a further shift to 400 nm). The protein binding pocket shifts the pKa of the chromophore, such that it is deprotonated at neutral pH. The remaining shift to 446 nm involves more subtle protein-chromophore interactions. The hydrogen bonds between the phenolic oxygen of the chromophore and active site side chains (Glu46 and Tyr42) are one important factor (Genick 1997). In addition, the position of Arg52, which may play a role as a counterion for the negative charge on the p-coumaric acid chromophore, and electronic polarization effects of the protein binding pocket have been invoked (Yoda 2001). However, significant open questions remain on the detailed physical mechanism involved in the spectral tuning effects of the PYP chromophore binding pocket.
The primary photochemical event that initiates the PYP photocycle has been identified as the isomerization of the central C=C double bond of the chromophore (Kort 1996a, 2004; Anderson 2004). Initially, it was thought that this isomerization event would result in a flip in the position of the aromatic ring of the chromophore, but subsequent FTIR (Fourier transform infared) experiments revealed that in fact it is the carbonyl group of the chromophore that undergoes the largest change in position during the first step of the PYP photocycle (Xie 1996; 2001)(Figure 5). Subsequent studies using X-ray crystallography at low temperature (Genick 1998; Kort 2004; Anderson 2004), time-resolved X-ray crystallography (Ren 2001), and time-resolved resonance Raman spectroscopy (Unno 2002) confirmed the motion of the carbonyl group, although the details of the chromophore motion are still under intense investigation. A key consequence of the isomerization mechanism, in which the position of the carbonyl flips, is that the overall motions of the chromophore are quite small, allowing the phenolic oxygen of the chromophore remain adjacent to active site residue Glu46 (see below).
The PYP photocycle shows a striking resemblance to that of the archaeal sensory rhodopsins (Meyer 1987; Hoff 1994). After the initial photoisomerization of the chromophore to form an early intermediate with a red-shifted absorbance spectrum (pR), the chromophore protonates, yielding a long-lived blue-shifted intermediate (pB). The pathway of proton transfer to the chromophore has been studied. FTIR spectroscopic data have led to a view in which a direct proton transfer occurs from active side chain Glu46 to the chromophore, yielding the intermediate pB' (Xie 1996, 2001; Imamoto 1997; Brudler 2001). As a result of this proton transfer, the side chain of Glu46, which is located in a hydrophobic pocket, becomes negatively charged. It has been proposed that the transfer of the buried negative charge from the chromophore (a stabilized, favorable buried charge) to Glu46 (an unfavorable buried charge) is a major part in driving the structural changes that occur during the pB' to pB transition (Xie 2001; Derix 2003). An alternative view is that chromophore protonation and Glu46 deprotonation are independent events, both driven by the strain caused by chromophore isomerization (Borucki 2002). The last step in the PYP photocycle is the recovery of the initial pG ground state from the pB intermediate. In view of its lifetime and its resemblance to the blue-shifted intermediates in the photocycles of the archaeal sensory rhodopsins, the pB intermediate is thought to be the signaling state of PYP.
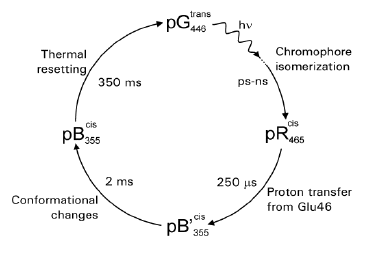
Figure 5. Schematic representation of the photocycle of photoactive yellow protein. Light-triggered transitions are indicated by wavy arrows, thermal transitions by straight arrows. Absorbance maxima are shown as subscripts. The time constants and main molecular processes for the transitions are indicated.
Because of it presumed functional role, the pB intermediate has been studied by a range of techniques. In particular, the nature of the structural changes that occur during pB formation have been studied, in order to understand how a receptor is converted to its active signaling state. Surprisingly, a number of results show that the pB state is a partially unfolded state, which shares properties with the molten globule state known from protein folding studies (Van Brederode 1996; Lee 2001a,b; Zhao 2006). This showed that folding and signaling are related in PYP. However, the structure of pB obtained by time-resolved X-ray crystallography does not reveal any unfolding (Genick 1997). This can be explained by the effect of the crystal lattice to reduce the structural changes that occur during the PYP photocycle (Xie 2001).
Striking analogies between the functioning of the SRs and PYP. The archaeal sensory rhodopsins and PYP have very different structures. The SRs are transmembrane
 -helical proteins with a retinal chromophore, while PYP is a water soluble protein that consists of a central
-helical proteins with a retinal chromophore, while PYP is a water soluble protein that consists of a central  -sheet, flanked on both sides by
-sheet, flanked on both sides by  -helices, and that has a p-coumaric acid chromophore. Despite these large differences in structure, striking analogies exist in the functioning of these two types of photoreceptors. Both are fund in extremely halophilic prokaryotes that live in hypersaline lakes, and both are involved in phototaxis. At the molecular level, both photoreceptors exhibit photocycles that are initiated by chromophore photoisomerization. In both cases, this initial event is followed by a proton transfer step that neutralizes the chromophore. And in both photoreceptors, this proton transfer step is a central part of the receptor activation mechanism. This observation confirms the central role of electrostatics in receptor activation. In the case of SRII, proton transfer results in the elimination of a key active site salt bridge, while in PYP it results in the formation of an unstable buried charge. Thus, these photoreceptors provide a remarkable case of convergent evolution, and their comparison allows the identification of general rules that govern photoreceptor activation.
-helices, and that has a p-coumaric acid chromophore. Despite these large differences in structure, striking analogies exist in the functioning of these two types of photoreceptors. Both are fund in extremely halophilic prokaryotes that live in hypersaline lakes, and both are involved in phototaxis. At the molecular level, both photoreceptors exhibit photocycles that are initiated by chromophore photoisomerization. In both cases, this initial event is followed by a proton transfer step that neutralizes the chromophore. And in both photoreceptors, this proton transfer step is a central part of the receptor activation mechanism. This observation confirms the central role of electrostatics in receptor activation. In the case of SRII, proton transfer results in the elimination of a key active site salt bridge, while in PYP it results in the formation of an unstable buried charge. Thus, these photoreceptors provide a remarkable case of convergent evolution, and their comparison allows the identification of general rules that govern photoreceptor activation.
iii. The BLUF photoreceptor PixD. The genome sequence of Synechocystis (Nakama 2000) contains a BLUF protein encoded by Slr1694 (later called PixD) (Gomelsky and Klug 2002). This protein has an absorbance maximum at 443 nm, with a second peak at 378 nm and shoulders at 363 and 471 nm (Masuda 2004), very similar to what has been reported for the AppA BLUF protein from Rhodobacter sphaeroides (Masuda and Bauer 2002). Illumination causes a characteristic ~10 nm redshift in the flavin absorbance maximum (to 457 nm). The recovery time of this red-shifted species is ~10 seconds (Masuda 2004; Okajima 2005), much shorter than the ~15 minute recovery time of AppA (Masuda and Bauer 2002). Disruption of this gene has the unexpected effect of altering the positive phototaxis response of Synechocystis towards 660 nm light into a negative phototaxis response (Okajima 2005), demonstrating that PixD is essential for the positive phototaxis response. The crystal structure of PixD has been reported at 1.8 Å resolution, revealing an
 /
/ fold and showing that the protein is stacked into two pentameric rings (Yuan 2006) (Figure 6).
fold and showing that the protein is stacked into two pentameric rings (Yuan 2006) (Figure 6).
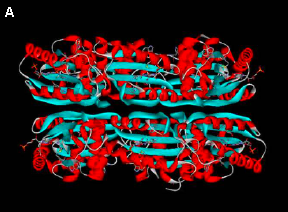
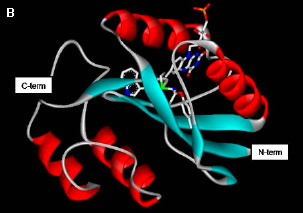
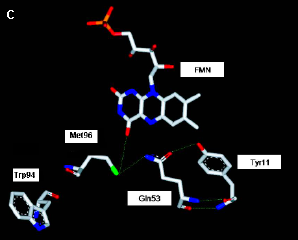
Figure 6. Structure of PixD. (A) Arrangement of PixD monomers into two stacked pentameric rings. (B) Cartoon representation of the structure of a PixD monomer, with-helices in red and
-sheets in blue. (C) The FAD chromophore is buried within the chromophore, and forms functionally important (see text and Figure 7) interactions with Tyr8 and Gln50. [Based on (Yuan 2006)]
The photochemistry of PixD has been studied by ultrafast spectroscopy (Gauden 2006). This provided evidence for a mechanism in which the excited state of the flavin undergoes light-induced electron transfer to Tyr8 on a picosecond time scale, followed by picosecond proton transfer from the Tyr to the positively charged flavin radical (Figure 7). This disrupts the hydrogen bond between the N5 of the flavin and Asn50, leaving this residue free to rotate by 180 degrees to form a novel hydrogen bond with the O4 of the flavin (Yuan 2006). This state is thought to be the signaling state and is fairly stable. It decays to the initial dark state in ~10 seconds.
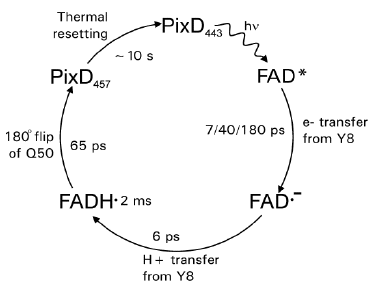
Figure 7. Schematic representation of the photocycle of PixD. Light-triggered transitions are indicated by wavy arrows, thermal transitions by straight arrows. Absorbance maxima are shown as subscripts. The time constants and main molecular processes for the transitions are indicated. Decay of the electronically excited FAD* state is multi-exponential. [Based on (Gauden 2006)]
iv. The cyanobacterial phytochrome pixJ1. The genome sequence of Synechocystis revealed multiple proteins related to phytochromes (Nakamura 2000). Phytochromes are photoreceptors that exhibit photochromism between two stable light-interconvertable species that usually have absorbance maxima in the red and far-red region. Their photochemistry is based on a linear tetrapyrrole chromophore that is attached to a Cys residue via a thioether (Rockwell 1996). Phytochromes consist of a multi-domain photosensory module and a signal transmitter module. In plant phytochromes the photosensory module is formed by three conserved domains: an N-terminal PAS domain, a central GAF domain, and a C-terminal PHY domain. In plant phytochromes the chromophore is attached to a Cys in the central GAF domain. However, bacterial phytochromes exhibit variations in this domain architecture. The chromophore attachment in bacteriophytochromes is to a Cys in the PAS domain close to the N-terminus. Cph2 from Synechocystis lacks the N-terminal domain, and in PixJ1 (see below) consists of two consecutive GAF domains followed by a transmitter domain homologous to methyl accepting chemotaxis proteins (MCP). The crystal structure for the PAS-GAF fragment of the bacteriophytochrome from Deinococcus radiodurans revealed an unusual knot in the N-terminal region (Wagner 2005). The crystal structure of the complete PAS-GAF-PHY sensory module of Chp1 from Synechocystis showed that the PAS domain forms an unusual tongue-like protrusion that seals the chromophore from the solvent (Essen 2008)(Figure 8).
Genetic disruption of PixJ1 (initially called Sll0041 and then PisJ1; also referred to as TaxD1) results in the loss of positive phototaxis in this organism (Yoshihara 2000; Bhaya 2001, 2004). Interestingly, this PixJ1 contains both a phytochrome domain and a domain related to methyl-accepting chemotaxis proteins (Yoshihara 2000; Bhaya 2001). PixJ1 exhibits a novel blue to green photoconversion, as opposed to the red to far-red photoconversion observed in other phytochromes (Yoshihara 2004, 2006), presumably triggered by photoisomerization over the C15-C16 double bond of its linear tetraypyrrole chromophore.
Mutants lacking the phytochrome Cph2 exhibit an aberrant response towards blue light (Wilde 2002; Friedler 2005), indicating that two different phytochromes are involved in the phototaxis responses of Synechocystis.
3. Signal Relay
Downstream signaling for the archaeal sensory rhodopsins. The sensory rhodopsins from H. salinarum are by far the best understood prokaryotic photosensory system with respect to their associated signal transduction chain, and thus form the model system for prokaryotic photosensory signal relay. Interestingly, the signal transduction chain of the SRs is quite similar to that involved in chemotaxis in E. coli.
A seminal finding in the field was the identification of the signal transducer for SRI as a methyl-accepting chemotaxis protein, named Htr for Halobacterial transducer protein, similar to the receptors responsible for chemotaxis in E. coli (Yao and Spudich 1992). This protein was first identified based on its reversible methylation during phototactic responses (Spudich 1989), and is located immediately upstream of the gene encoding the apoprotein of SRI (Yao and Spudich 1992). The Htr transducer proteins consist of two transmembrane
 -helices and a large intracellular domain that is active in downstream signaling to kinases (see below), and is methylated during signal adaptation, as is the case in E. coli chemotaxis. SRI and SRII each interact with their own associated transducer protein (HtrI and HtrII). The Hrt proteins form a tight and specific complex with their SR partners, and result in a tetrameric signaling complex consisting of two SR proteins and two Htr proteins (Chen and Spudich 2002). Photoexcitation of the SR-Htr complex alters interactions within the complex, such that a signal is relayed from the SR to the Htr. The molecular mechanism of this inter-protein signal relay has attracted significant attention, but at this point has not yet been fully resolved. A striking observation is that some mutations in SRI and HtrI result in an inverted signaling phenotype, where normally attractant orange light causes a repellent response (Olsen 1995; Jung and Spudich 1996). This phenomenon was recently explained based on a conformational changes in the SR/Htr complex that switches the connectivity of the Schiff base between the cytoplasm and the periplasm (Sineshchekov 2008). Mutants that alter the equilibrium between these two states cause inverted signaling.
-helices and a large intracellular domain that is active in downstream signaling to kinases (see below), and is methylated during signal adaptation, as is the case in E. coli chemotaxis. SRI and SRII each interact with their own associated transducer protein (HtrI and HtrII). The Hrt proteins form a tight and specific complex with their SR partners, and result in a tetrameric signaling complex consisting of two SR proteins and two Htr proteins (Chen and Spudich 2002). Photoexcitation of the SR-Htr complex alters interactions within the complex, such that a signal is relayed from the SR to the Htr. The molecular mechanism of this inter-protein signal relay has attracted significant attention, but at this point has not yet been fully resolved. A striking observation is that some mutations in SRI and HtrI result in an inverted signaling phenotype, where normally attractant orange light causes a repellent response (Olsen 1995; Jung and Spudich 1996). This phenomenon was recently explained based on a conformational changes in the SR/Htr complex that switches the connectivity of the Schiff base between the cytoplasm and the periplasm (Sineshchekov 2008). Mutants that alter the equilibrium between these two states cause inverted signaling.
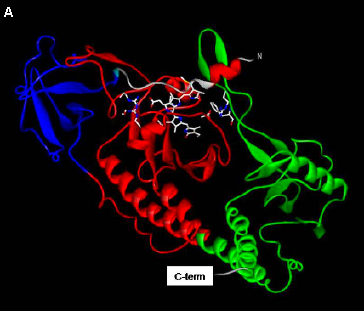
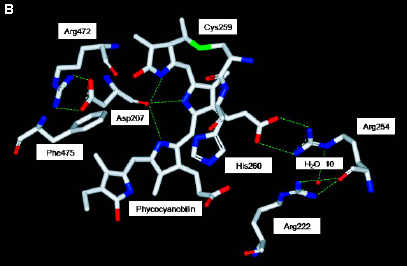
Figure 8. Structure of Cph1.
(A) Cartoon representation of the structure of Cph1. The PAS domain is indicated in red, the GAF domain in blue, and the Phy domain in green.
(B) The phycocyanobilin chromophore is fully enclosed by the PAS, GAF, and Phy domains. [Based on (Essen 2008)]
The binding of HtrI to SRI has a number of effects on the photocycle of SRI. First, the kinetics and the pH dependence of the photocycle kinetics are altered (Spudich and Spudich 1993), and the pathway for proton transfer is altered (Rath 1996). Secondly, in the absence of HtrI the SRI photoreceptor has transmembrane proton pumping activity (Bogomolni et al., 1994). In the presence of HtrI, this proton pumping activity is fully repressed. This suggests that the protein conformational change involved in transmembrane proton pumping is similar to that involved for signal relay from SRI to HtrI.
Significant progress has been made regarding the structural basis of the signal relay process from SRII from Natronobacterium to its transducer. First, an approach based on site-specific (Cys-directed) labeling of sites in the SRII/HtrII complex was followed, leading to a proposed outward movement of the F upon photoexcitation of SRII, in turn causing a rotation of TM2 in the transducer (Wegener 2000). Secondly, the crystal structure of SRII was determined, revealing that Tyr199 is exposed on the outside of the 7-helical bundle of the photoreceptor, close to the center of the lipid bilayer (Luecke 2001). Thus, this residue is an excellent candidate for interacting with the transducer protein. Third, the crystal structure of a complex containing SRII and the transmembrane segment of HtrII was reported (Gordeliy 2002). This structure confirmed a role for Tyr199 in interactions between SRII and its transducer, and provided detailed information on this complex.
In line with the similarity of the Htr proteins to the E. coli chemotaxis receptors, the signal transduction chain downstream of Htr is centered abound CheA and CheY proteins (Rudolph and Oesterhelt 1995, 1996). While each of the two SRs is associated with a dedicated Htr protein, their signaling chains converge at the level of CheA.
Downstream signaling for photoactive yellow protein. The signal transduction chain associated with the PYP from H. halophila has not yet been identified, largely because no genetic tools are available for this organism. A homolog of PYP has been identified in the genetically accessible bacterium Rhodobacter sphaeroides, but the puzzling observation was made that deletion of the pyp gene in this organism has no effect on its negative phototaxis response towards blue light (Kort 2000). The PYP homolog from R. centenaria did provide important insights into the downstream signal transduction pathway, because it is part of a much larger protein that also contains domains with clear homology to phytochromes and to sensor histidine kinases. The full-length protein exhibits light-induced changes in autophosphorylation activity, providing strong support for the notion that downstream signaling of the R. centenaria PYP is based on phosphorelay by a two-component regulatory system. This system was reported to regulate the gene expression of a chalcone synthetase. At this point it is not know if the PYP from H. halophila also operates via a sensor histidine kinase.
Phototactic signaling in Rhodocista centenaria. The purple photosynthetic bacterium Rhodocista centenaria exhibits positive phototaxis towards light <800 nm absorbed by the photosynthetic machinery and negative phototaxis towards light <600 nm (Ragatz 1995). The likely photoreceptor for the positive phototaxis response is photosynthetic machinery (see Section 2a). Note that the signal transduction chain for this response is largely unknown. The photoreceptor for the negative phototactic response has not yet been identified. However, using genetic approaches based on phototactic mutants some information has been obtained on the signal transduction chains in these two photoresponses in R. centenaria Rsp. centenum (Jiang 1997, 1998).
Analysis of mutants with perturbed phototactic responses resulted in the identification of a chemotaxis operon (Jiang 1998). Disruption of this operon has affects not only on chemotaxis responses, but also on phototaxis (Jiang 1997). Thus, phototaxis and chemotaxis share a common downstream signal transduction chain. Interestingly, both the positive and negative phototaxis responses were abolished in these mutants, suggesting that signaling of the redox sensor-based positive phototaxis response (see Section 2a) occurs via the chemotactic signal transduction chain.
Photosignaling in Synechocystis twitching motility. Yeast two-hybrid screens identified PixE as the interaction partner of PixD (Okajima 2005), and subsequent biochemical experiments confirmed this interaction (Yuan and Bauer 2008). In the dark a PixD10-PixE5 complex is formed, which is disrupted upon illumination (Yuan and Bauer 2008). This dark complex corresponds well with the funding that in protein crystals PixD forms two stacked pentameric rings (Yuan 2006). Illumination results in the release of PixE monomers and PixD dimers, which presumably interact with downstream proteins. A separate series of studies has shown a role of cAMP in the phototaxis responses of Synechocystis (Ohmori and Okamoto 2004). Mutants in the adenylate cyclase Cya1 exhibit perturbed motility, which was restored by the addition of exogenous cAMP (Terauchi and Ohmiro 1999, 2004; Bhaya 2006). In addition, blue light stimuli result in elevated cAMP levels through the activity of Cya1 (Masuda and Ono 2004). Large-scale screening of protein-protein interactions (Sato 2007) has revealed interactions between PixD and glucokinase and glycogen phosphorylase. Thus, light-induced changes in glycogen metabolism may be the cause of the observed changes in cAMP (Hoff 2009). The binding of cAMP to the cAMP receptor protein SYCRP1 regulates the activity of this protein as a transcription factor for the biogenesis of pili (Yoshimura 2002). It remains to be seen if this pathway also is responsible for phototaxis responses.
4. Changes in Motility
Prokaryotic phototaxis involves a change in the motility of the bacterium induced by changes in the light climate that the bacterium experiences. An important distinction in describing phototactic responses is responses that are based on the intensity of light, in which the cell compares the current light intensity with that of a short time ago, and responses in which the actual direction of light is measured. Responses in which the intensity, but not the direction of light is measured, are usually referred to as photophobic responses, while responses that are based on the actual direction of the light stimulus are considered true phototaxis responses.
The phototaxis responses in H. salinarum, Rb. sphaeroides, and H. halophila described above occur in free-swimming prokaryotes. This type of motility is based on the rotary bacterial flagellum. A significant degree of variability exist regarding the morphology of the flagellum and the swimming behavior of the bacterium. Rb. sphaeroides swims by means of a single flagellum located on the side of the cell. H. halophila has two flagella, one on each of the tips of the cell. H. salinarum uses tufts of flagella located on the cell's poles. All of these flagellar morphologies are distinct from the bundle of flagella that E. coli uses for chemotaxis.
The type of motility that prokaryotes display in phototactic responses is also diverse. Prokaryotes can be free-swimming in solution, using their rotary flagella. This type of motion has been studied most extensively. In addition, bacteria can swarm over a solid medium. A relevant example is provided by R. centenaria, which can swim in liquid medium by means of a single polar flagellum, but can also swarm over a solid surface using large numbers of lateral flagella. Interestingly, entire colonies of R. centenaria can move over a solid surface (Ragatz 1995). Finally, some bacteria, particularly cyanobacteria, exhibit sliding motility on solid surfaces (Bhaya 2000; McBride 2001).
Free-swimming prokaryotes perform phototaxis by changing the rotation direction of their flagella upon exposure to light. Two typical photoresponses are inversion of swimming direction, as observed in H. salinarum and H. halophila, and a transient stop of rotation of the flagellum, as displayed by Rb. sphaeroides. In addition, Rb. sphaeroides displays photokinesis, in which the speed of swimming is modulated by light stimuli.
A step-up or step-down stimulus evokes a response in the cell population under study that consists of two phases. First, during the excitation phase a rapid change in the frequency of altering the rotation of the flagellar motor is observed. This phase typically occurs on a time-scale of a 1 second. Secondly, an approximately 10-fold slower adaptation process takes place, which resets the behavior of the flagellum to the pre-stimulus value, even in the continued presence of the light stimulus. The excitation phase is caused by the formation of the receptor signaling state, and amplification of its signal by the downstream signal transduction chain. The adaptation phase in H. salinarum phototaxis (and E. coli chemotaxis) is caused by methylation of the Htr transducer proteins.
All responses studied in free-swimming prokaryotes are photophobic or photokinetic. Thus, it was thought that prokaryotes are not able to respond to the direction of light. However, the movement of entire colonies of R. centenaria appears to be guided by the actual direction of the light, and thus constitutes a true phototaxis response (Ragatz 1995; Gest 1995).
Synechocystis exhibits complex photomovement behavior (Yoshihara and Ikeuchi, 2004), with non-linear dose-response curves, involving multiple light inputs (Ng 2003; Montgomery 2007), as described above: PixD, PixJ1, and Cph2 It shows positive phototaxis towards light in the spectral region 560 - 720 nm, and negative phototaxis towards 260 nm light (Ng 2003; Choi 1999). At high light intensities, 470 nm light and light in the region 600-700 nm also induce negative phototaxis. The movement of Synechocystis is based on the synthesis and retraction of Typi IV pilli (Bhaya 2000).
Acknowledgements: The figures depicting the photoreceptor crystal structures were prepared by Dr. Masato Kumauchi.
REFERENCES
Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer, C. 2005. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry 44, 7998-8005.
Anderson S, Srajer V, Moffat K. 2004. Structural heterogeneity of cryotrapped intermediates in the bacterial blue light photoreceptor, photoactive yellow protein, Photochemistry and Photobiology 80, 7-14.
Armitage JP, Hellingwerf KJ. 2003. Light-induced behavioral responses ('phototaxis') in prokaryotes. Photosynth. Res. 76, 145-155.
Armitage JP. 1997. Behavioural responses of bacteria to light and oxygen. Arch. Microbiol. 168: 249-261.
Baca M, Borgstahl GEO, Boissinot M, Burke PM, Williams DR, Slater KA, Getzoff ED. 1994. Complete chemical structure of photoactive yellow protein: Novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry 33, 14369-14377.
Berry RM, Armitage JP. 2000. Response kinetics of tethered Rhodobacter sphaeroides to changes in light intensity. Biophys. J. 78, 1207-1215.
Bhaya D, Bianco NR, Bryant D, Grossman A. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37, 941-951.
Bhaya D, Nakasugi K, Fazeli F, Burriesci MS. 2006. Phototaxis and impaired motility in adenylyl cyclase and cyclase receptor protein mutants of Synechocystis sp. strain PCC 6803. J. Bacteriol. 188, 7306-7310.
Bhaya D, Takahashi A, Grossman AR. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98, 7540-7545.
Bhaya, D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Micro. 53, 745-754.
Bibikov SI, Grishanin RN, Kaulen AD, Marwan W, Oesterhelt D, Skulachev VP. 1993. Bacteriorhodopsin is involved in halobacterial photoreception. Proc. Natl. Acad. Sci. USA 90, 9446-9450.
Birge RR, Cooper TM. 1983. Energy storage in the primary step of tf the photocycle of bacteriorhodopsin. Biophys. J. 42, 61-69.
Borgstahl GEO, Williams DR, Getzoff ED. 1995. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site and chromophore. Biochemistry 34, 6278-6287.
Borucki B, Devanathan S, Otto H, Cusanovich MA, Tollin G, Heyn MP. 2002. Kinetics of proton uptake and dye binding by photoactive yellow protein in wild type and in the E46Q and E46A mutants. Biochemistry 41, 10026-10037.
Briggs WR, Christie JM, Salomon M. 2001. Phototropins: A new family of flavin-binding blue light receptors in plants. Antioxid. Redox Signal. 3, 775-788.
Briggs WR. 2007. The LOV domain: a chromophore module servicing multiple photoreceptors. J. Biomed. Sci. 14, 499-504.
Brudler R, Rammelsberg R, Woo TT, Getzoff ED, Gerwert K. 2001. Structure of the I-1 early intermediate of photoactive yellow protein by FTIR spectroscopy. Nat. Struct. Biol. 8, 265-270.
Chen XP, Spudich, JL. 2002. Demonstration of 2 : 2 stoichiometry in the functional SRI-HtrI signaling complex in Halobacterium membranes by gene fusion analysis. Biochemistry 41, 3891-3896.
Choi JS, Chung YH, Moon YJ, Kim C, Watanabe M, Song PS, Joe CO, Bogorad L, Park YM. 1999. Photomovement of the gliding cyanobacterium Synechocystic sp. PCC 6803. Photochem. Photobiol. 70, 95-102.
Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96, 8779-8783.
Crosson S, Rajagopal S, Moffat K. 2003. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42, 2-10.
Cusanovich MA, Meyer TE. 2003. Photoactive yellow protein: A prototypic PAS domain sensory protein and development of a common signaling mechanism. Biochemistry 42, 4759-4770.
Davis SJ, Vener AV, Vierstra RD. 1999. Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286, 2517-2520.
Derix NM, Wechselberger RW, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. 2003. Lack of negative charge in the E46Q mutant of photoactive yellow protein prevents partial unfolding of the blue-shifted intermediate. Biochemistry 42, 14501-14506.
Dux P, Rubinstenn G, Vuister GW, Boelens R. Mulder AA, Hard K, Hoff WD, Kroon AR, Crielaard W, Hellingwerf KJ, Kaptein R. 1998. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry 37, 12689-12699.
Engelhard M, Scharf B, Siebert F. 1996. Protonation changes during the photocycle of sensory rhodopsin II from Natronobacterium pharaonis. FEBS Lett. 395, 195-198.
Essen O-L, Mailliet J, Hughes J. 2008. The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. USA 105, 14709-14714.
Fiedler B, Borner T, Wilde A. 2005. Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803: Role of different photoreceptors. Photochem. Photobiol. 81, 1481-1488.
Flower DR (1999) Modelling G-protein-coupled receptors for drug design. Biochim Biophys Acta 1422: 207-234.
Gauden, M, van Stokkum, IHM, Key, JM, Luhrs, DC, Van Grondelle, R, Hegemann, P, Kennis, JTM. 2006. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. USA 103, 10895-10900.
Genick UK, Borgstahl GE, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED. 1997. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science 275, 1471-1475.
Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. 1997. Active site mutants implicate key residues for control of color and light cycle kinetics of photoactive yellow protein. Biochemistry 36, 8-14.
Genick UK, Soltis SM, Kuhn P, Canestrelli IL, Getzoff ED. 1998. Structure at 0.85 Å resolution of an early protein cycle intermediate. Nature 392, 206-209.
Gest H. 1995. Phototaxis and other sensory phenomena in purple photosynthetic bacteria. FEMS Microbiol. Lett. 16, 287-294.
Getzoff ED, Gutwin KN, Genick UK. 2003. Anticipatory active-site motions and chromophore distortion prime photoreceptor PYP for light activation. Nat. Struct. Biol. 10, 663-668.
Gomelsky M, Kaplan S. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 273, 35319-35325.
Gomelsky M, Klug G. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27: 497-500.
Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. 2002. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 419, 484-487.
Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. 1996. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259, 393-421.
Grishanin RN, Bibikov SI, Altschuler IM, Kaulen AD, Kazimirchuk SB, Armitage JP, Skulachev VP. 1996. Delta psi-mediated signalling in the bacteriorhodopsin-dependent photoresponse. J. Bacteriol. 178, 3008-3014.
Grishanin RN, Gauden DE, Armitage JP. 1997. Photoresponses in Rhodobacter sphaeroides: Role of photosynthetic electron transport. J. Bacteriol. 179, 24-30.
Gudermann, B. Nurnberg and G. Schultz, Receptors and G-proteins as primary components of transmembrane signal transduction. J. Mol. Med. 73 (1995), pp. 51-63.
Hader DP. 1987. Photosenory behavior in prokaryotes. Microbiol. Rev. 51, 1-21.
Hayashi S, Tajkhorshid E, Pebay-Peyroula E, Royant A, Landau EM, Navarro J, Schulten K. 2001. Structural determinants of spectral tuning in retinal proteins-bacteriorhodopsin vs sensory rhodopsin II. J. Phys. Chem. 105, 10124-10131.
Hellingwerf KJ, Hendriks J, Gensch T. 2003. Photoactive Yellow Protein, a new type of photoreceptor protein: Will this "yellow lab" bring us where we want to go? J. Phys. Chem. 107, 1082-1094.
Hoff WD, Devreese B, Fokkens R, Nugteren-Roodzant IM, Van Beeumen J, Nibbering N, Hellingwerf KJ. 1996. Chemical reactivity and spectroscopy of the thiol ester-linked p-coumaric acid chromophore in the photoactive yellow protein from Ectothiorhodospira halophila. Biochemistry 35, 1274-1281.
Hoff WD, Dux P, Hard K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens, R, Van Beeumen J, Hellingwerf KJ. 1994. Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry 33, 13959-13962.
Hoff WD, Jung KH, Spudich JL. 1997. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 26, 223-258.
Hoff WD, van der Horst MA, Nudel CB, Hellingwerf KJ. 2009. Prokaryotic phototaxis. In Methods in Molecular Biology: chemotaxis (Humana Press), in press.
Hu JG, Griffin RG, Herzfeld J. 1994. Synergy in the spectral tuning of retinal proteins - complete accounting of the opsin shift in baceriorhodopsin. Proc. Natl. Acad. Sci. USA 91, 8880-8884.
Hustede E, Liebergesell M, Schlegel HG. 1980. The photophobic response of various sulfur and nonsulfur purple bacteria. Photochem. Photobiol. 50, 809-815.
Imamoto Y, Ito Y, Kataoka M, Tokunaga, F. 1995. Reconstitution of photoactive yellow protein from apoprotein and p-coumaric acid derivatives. FEBS Lett. 374, 157-160.
Imamoto Y, Kataoka M. 2007. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem. Photobiol. 83, 40-49.
Imamoto Y, Mihara K, Hisatomi O, Kataoka M, Tokunaga F, Bojkova N, Yoshihara K. 1997. Evidence for proton transfer from Glu-46 to the chromophore during the photocycle of photoactive yellow protein. J. Biol. Chem. 272, 12905-12908.
Jiang Z, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE. 1999. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285, 406-409.
Jiang ZY, Gest H, Bauer CE. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 179, 5720-5727.
Jiang ZY, Rushing BG, Bai Y, Gest H, Bauer CE. 1998. Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J. Bacteriol. 180, 1248-1255.
Jung A, Domratcheva T, Tarutina M, Wu Q, Ko WH, Shoeman RL, Gomelsky M, Gardner KH, Schlichting I. 2005. Structure of a bacterial BLUF photoreceptor: Insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 102, 12350-12355.
Jung, KH, Spudich, JL. 1996. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc. Natl. Acad. Sci. USA 93, 6557-6561.
Kennis JTM, Groot ML. 2007. Ultrafast spectroscopy of biological photoreceptors. Curr. Opin. Struct. Biol. 17, 623-630.
Kita, A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. 2005. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J. Mol. Biol. 349, 1-9.
Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. 1999. How color visual pigments are tuned. Trends Biochem. Sci. 24, 300-305.
Kolbe M, Besir H, Essen LO, Oesterhelt D. 2000. Structure of the light-driven chloride pump halorhodopsin at 1.8 angstrom resolution. Science 288, 1390-1396.
Kort R, Crielaard W, Spudich JL, Hellingwerf KJ. 2000. Color-sensitive motility and methanol release responses in Rhodobacter sphaeroides, Journal of Bacteriology 182, 3017-3021.
Kort R, Hellingwerf KJ, Ravelli RBG. 2004. Initial events in the photocycle of photoactive yellow protein, Journal of Biological Chemistry 279, 26417-26424.
Kort R, Hoff WD, Van West M, Kroon AR, Hoffer SM., Vlieg KH, Crielaard W, Van Beeumen JJ, Hellingwerf KJ. 1996b. The xanthopsins: A new family of eubacterial blue-light photoreceptors, EMBO J. 15, 3209-3218.
Kort R, Vonk H, Xu X, Hoff WD, Crielaard W, Hellingwerf KJ. 1996a. Evidence for the trans-cis isomerization of the p-coumaric acid chromophore as the photochemical basis of the photocycle of photoactive yellow protein. FEBS Lett. 382, 73-78.
Kroon A, Hoff WD, Fennema H, Koomen G-J, Verhoeven JW, Crielaard W, Hellingwerf, KJ. 1996. Spectral tuning, fluorescence, and photoactivity in hybrids of photoactive yellow protein, reconstituted with native or modified chromophores. J. Biol. Chem. 271, 31949-31956.
Kumauchi M, Hara M, Stalcup P, Xie A, Hoff WD. 2008. Identification of six new photoactive yellow proteins: diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem. Photobiol. 84, 956-969.
Lee B-C, Croonquist PA, Sosnick, TR, Hoff WD. 2001a. PAS domain receptor photoactive yellow protein is converted to a molten globule state upon activation. J. Biol. Chem. 276, 20821-20823.
Lee B-C, Pandit A, Croonquist PA, Hoff WD. 2001b. Folding and signaling share the same pathway in a photoreceptor. Proc. Natl. Acad. Sci. USA 98, 9062-9067.
Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. 2001. Crystal structure of sensory rhodopsin II at 2.4 angstroms: Insights into color tuning and transducer interaction. Science 293, 1499-1503.
Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. 1999. Structure of bacteriorhodopsin at 1.55 angstrom resolution. J. Mol. Biol. 291, 899-911.
Maeda A, Sasaki J, Yamazaki Y, Needleman R, Lanyi JK. 1994. Interaction of aspartate 85 with a water molecule and the protonated Schiff base in the L intermediate of bacteriorohodpsin - a Fourier transform infrared spectroscopic study. Biochemistry 33, 1713-1717.
Marti T, Rosselet SJ, Otto H, Heyn MP, Khorana HG. 1991. The retinylidene Schiff base counterion in bacteriorhodopsin. J. Biol. Chem. 266, 18674-18683.
Marwan W, Oesterhelt D. 1990. Quantitation of photochromism of sensory rhodopsin I by computerized tracking of Halobacterium halobium cells. J. Mol. Biol. 215, 277-285.
Masuda S, Bauer CE. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110, 613-623.
Masuda S, Hasegawa K, Ishii A, Ono T. 2004. Light-induced structural changes in a putative blue-light receptor with novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of Synechocystis sp. PCC6803. Biochemistry 43, 5304-5313.
Masuda, S, Ono T. 2004, Biochemical characterization of the major adenylyl cyclase, Cya1, in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 577, 255-258.
McBride MJ. 2001. Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55, 49-75.
Memmi S, Kyndt J, Meyer T, Devreese B, Cusanovich M, Van Beeumen J. 2008. Photoactive yellow protein from the halophilic bacterium Salinibacter ruber. Biochemistry 47, 2014-2024.
Meyer TE, Yakali E, Cusanovich, MA, Tollin G. 1987. Properties of a water soluble, yellow protein isolated from a halophilic phototrophic bacterium that has photochemical activity analogous to sensory rhodopsin. Biochemistry 26, 418-423.
Montgomery BL. 2007. Sensing the light: photoreceptive systems and signal transduction in cyanobacteria. Mol. Micro. 64, 16-27.
Nakamura Y, Kaneko T, Tabata S. 2000. CyanoBase, the genome database for Synechocystis sp. strain PCC6803: status for the year 2000. Nucleic Acids Res. 28, 72.
Ng WO, Grossman AR, Bhaya D. 2003. Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803. J. Bacteriol. 185, 1599-1607.
Ohmori M, Okamoto S. 2004. Photoresponsive cAMP signal transduction in cyanobacteria. Photochem. Photobiol. Sci. 3, 503-511.
Okajima S, Yoshihara S, Fukushima Y, Geng X, Katayama M, Higashi S, Watanabe M, Sato S, Tabata S, Shibata Y, Itoh S, Ikeuchi M. 2005. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J. Biochem. 137, 741-750.
Olsen KD, Zhang XN, Spudich JL. 1995. Residue replacements of buried aspartyl and related residues in sensory rhodopsin - D201N produces inverted phototaxis signals. Proc. Natl. Acad. Sci. USA 92, 3185-3189.
Ragatz L, Jiang ZY, Bauer CE, Gest H. 1994. Phototactic purple bacteria. Nature 370, 104-104.
Ragatz L, Jiang ZY, Bauer CE, Gest H. 1995. Macroscopic phototactic behavior of the purple photosynthetic bacterium Rhodospirillum centenum. Arch. Microbiol. 163, 1-6.
Rajagopal S, Moffat K. 2003. Crystal structure of a photoactive yellow protein from a sensor histidine kinase: Conformational variability and signal transduction. Proc. Natl. Acad. Sci. USA 100, 1649-1654.
Rath P, Olsen KD, Spudich JL, Rothschild KJ. 1994. The Schiff base counterion of bacteriorhodopsin is protonated in sensory rhodopsin I. Spectroscopic and functional characterization of the mutated proteins D76A and D76A. Biochemistry 33, 5600-5606
Rath, P, Spudich, E, Neal DD, Spudich, JL, Rothschild, KJ. 1996. Asp76 is the Schiff base counterion and proton acceptor in the proton-translocating form of sensory rhodopsin I. Biochemistry 35, 6690-6696.
Ren L, Martin CH, Wise KJ, Gillespie NB, Luecke H, Lanyi JK, Spudich JL, Birge RR. 2001. Molecular mechanism of spectral tuning in sensory rhodopsin II. Biochemistry 40, 13906-13914.
Ren Z, Perman B, Srajer V, Teng TY, Pradervand C, Bourgeois D, Schotte F, Ursby T, Kort R, Wulff M, Moffat K. 2001. A molecular movie at 1.8 angstrom resolution displays the photocycle of photoactive yellow protein, a eubacterial blue-light receptor, from nanoseconds to seconds. Biochemistry 40, 13788-13801.
Rockwell NC, Su YS, Lagarias JC. 2006. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant. Biol. 57, 837-858.
Royant A, Nollert P, Edman K, Neutze R, Landau EM, Pebay-Peyroula E, Navarro J. 2001. X-ray structure of sensory rhodopsin II at 2.1-angstrom resolution. Proc. Natl. Acad. Sci. USA 98, 10131-10136.
Rudolph J, Oesterhelt D. 1995. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarum. EMBO J. 14, 667-673.
Rudolph J, Oesterhelt D. 1996. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J. Mol. Biol. 258, 548-554.
Sackett MJ, Armitage JP, Sherwood EE, Pitta TP. 1997. Photoresponses of the purple nonsulfur bacteria Rhodospirillum centenum and Rhodobacter sphaeroides. J. Bacteriol. 179, 6764-6768.
Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Reviews 103, 2203-2237.
Sasaki J, Spudich JL. 2008. Signal transfer in haloarchaeal sensory rhodopsin-transducer complexes. Photochem. Photobiol. 84, 863-868.
Sato S, Shimoda Y, Muraki A, Kohara M, Nakamura Y, Tabata S. 2007. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 14, 207-216.
Sgarbossa A, Checcucci G, Lenci F. 2002. Photoreception and photomovements of microorganisms. Photochem. Photobiol. Sci. 1, 459-467.
Sineshchekov OA, Sasaki J, Phillips BJ, Spudich JL. 2008. A Schiff base connectivity switch in sensory rhodopsin signaling. Proc. Natl. Acad. Sci. USA 105, 16159-16164.
Sprenger WW, Hoff WD, Armitage JP, Hellingwerf KJ. 1993. The Eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorbance spectrum of the photoactive yellow protein. J. Bacteriol. 175, 3096-3104.
Spudich EN, Spudich JL. 1993. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J. Biol. Chem. 268, 16095-16097.
Spudich EN, Takahashi T, Spudich JL. 1989. Sensory rhodopsin I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc. Natl. Acad. Sci. USA 86, 7746-7750.
Spudich EN, Zhang WS, Alam M, Spudich JL. 1997. Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base Asp-73 interhelical salt bridge. Proc. Natl. Acad. Sci. USA 94, 4960-4965.
Spudich JL, Bogomolni RA. 1984. Mechanism of color discrimination by a bacterial sensory rhodopsin. Nature 312, 509-513.
Spudich JL, Bogomolni RA. 1988. Sensory rhodopsins of halobacteria. Annu. Rev. Biophys. Biophys. Chem. 17, 193-215.
Sudo Y, Spudich JL. 2006. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc. Natl. acad. Sci. USA 103, 16129-16134.
Taylor BL, Zhulin IB. 1999. PAS domains: Internal sensors of oxygen, redox potential, and light, Microbiol. Mol. Biol. Rev. 63, 479-506.
Terauchi K, Ohmori M. 1999. An adenylate cyclase, Cya1, regulates cell motility in the cyanobacterium Synechocystis sp. PCC 6803. Plant. Cell. Physiol. 40, 248-251.
Terauchi K, Ohmori M. 2004. Blue light stimulates cyanobacterial motility via a cAMP signal transduction system. Mol. Micro. 52, 303-309.
Unno M, Kumauchi M, Sasaki J, Tokunaga F, Yamauchi S. 2002. Resonance Raman spectroscopy and quantum chemical calculations reveal structural changes in the active site of photoactive yellow protein. Biochemistry 41, 5668-5674.
Van Beeumen J, Devreese B, Van Bun S, Hoff WD, Hellingwerf KJ, Meyer TE, McRee, DE, Cusanovich MA. 1993. The primary structure of a photoactive yellow protein from the phototrophic bacterium, Ectothiorhodospira halophila, with evidence for the mass and the binding site of the chromophore. Protein Science 2, 1114-1125.
Van Brederode ME, Hoff WD, Van Stokkum IHM, Groot ML, Hellingwerf KJ. 1996. Protein folding thermodynamics applied to the photocycle of the photoactive yellow protein. Biophys. J. 71, 365-380.
Van der Horst, M.A., Hellingwerf, K.J. 2004. Photoreceptor proteins, "star actors of modern times": A review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37, 13-12.
Wagner, JR; Brunzelle, JS; Forest, KT; Vierstra, RD. 2005. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325-331.
Wegener, AA, Chizhov, I, Engelhard, M, Steinhoff, HJ. 2000. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J. Biol. Chem. 301, 881-891.
Wilde A, Fiedler B, Borner T. 2002. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Micro. 44, 981-988.
Xie A, Hoff WD, Kroon AR, Hellingwerf KJ. 1996. Glu46 donates a proton to the 4-hydroxycinnamate anion chromophore during the photocycle of photoactive yellow protein. Biochemistry 35, 14671-14678.
Xie A, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. 2001. Formation of a new buried charge drives a large-amplitude protein quake in photoreceptor activation. Biochemistry 40, 1510-1517.
Yan B, Spudich JL, Mazur P, Vunnam S, Derguini F, Nakanish K. 1995. Spectral tuning in bacteriorhodopsin in the absence of counterion and coplanarization effects. J. Biol. Chem. 270, 29668-29670.
Yan B, Spudich JL. 1991. Evidence that the repellent receptor form of sensory rhodopsin I is an attractant signaling state. Photochem. Photobiol. 54, 1023-1026.
Yan B, Takahashi T, Johnson R, Spudich JL. 1991. Identification of signaling states of a sensory receptor by modulation of lifetimes of stimulus-induced conformations - the case of sensory rhodopsin II. Biochemistry 30, 10686-10692.
Yao, VJ, Spudich JL. 1992. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc. Natl. Acad. Sci. USA 89, 11915-11919.
Yoda M, Houjou H, Inoue Y, Sakurai M. 2001. Spectral tuning of photoactive yellow protein. Theoretical and experimental analysis of medium effects on the absorption spectrum of the chromophore J. Phys. Chem. 105, 9887-9895.
Yoshihara S, Ikeuchi M. 2004. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 3, 512-518.
Yoshihara S, Katayama M, Geng X, Ikeuchi M. 2004. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 45, 1729-1737.
Yoshihara S, Shimada T, Matsuoka D, Zikihara K, Kohchi T, Tokutomi S. 2006. Reconstitution of blue-green reversible photoconversion of a cyanobacterial photoreceptor, PixJ1, in phycocyanobilin-producing Escherichia coli. Biochemistry 45, 3775-3784.
Yoshihara S, Suzuki F, Fujita H, Geng XX, Ikeuchi M. 2000. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 41, 1299-1304.
Yoshihara, S, Katayama, M, Geng, XX, Ikeuchi, M. 2004. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 45, 1729-1737.
Yoshimura H, Yoshihara S, Okamoto S, Ikeuchi M, Ohmori M. 2002. A cAMP receptor protein, SYCRP1, is responsible for the cell motility of Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 460-463.
Yuan H, Anderson S, Masuda S, Dragnea V, Moffat K, Bauer, C. 2006. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry 45, 12687-12694.
Yuan H, Bauer CE. 2008. PixE promotes dark oligomerization of the BLUF photoreceptor PixD. Proc. Natl. Acad. Sci. USA 105, 11715-11719.
Zhang XN, Spudich JL. 1997. His166 is critical for active-site proton transfer and phototaxis signaling by sensory rhodopsin I. Biophys. J. 73, 1516-1523.
Zhao JM, Lee H, Nome RA, Majid S, Scherer NF, Hoff WD. 2006. Single-molecule detection of structural changes during PAS domain activation. Proc. Natl. Acad. Sci. USA. 103, 1561-11566.
2/3/09