BLUE LIGHT SENSING in PLANTS
All You Need is a Little LOV
John M. Christie
Division of Molecular and Cellular Biology
Faculty of Biomedical and Life Sciences
University of Glasgow,
Glasgow G12 8QQ, UK
J.Christie@bio.gla.ac.uk
Blue Light And Phototropism
Unlike humans, plants are immobile, and at the mercy of their surrounding environment. Consequently, they have evolved sophisticated mechanisms that enable them to perceive and respond to environmental changes, and to modify their growth accordingly. Some of these processes actually involve movement. Not movement of the plant itself, but movement of various parts of the plant. One example is phototropism, a Greek term describing the process by which plants grow towards light.
As early as 1880, Charles Darwin had noted: "no one can look at the plants growing on a bank or on the borders of a thick wood, and doubt that the young stems and leaves place themselves so that the leaves may be well illuminated" (1). Phototropism is especially important for germinating seedlings, whereby the emerging shoot must grow towards the light to survive by maximizing the capture of light for photosynthesis (Figure 1).

Figure 1. Time-lapse image series showing the phototropism response of an oat seedling. After 3 days of growth in the dark, the oat seedling was exposed to dim blue light from the left hand side for 9 hours. Pictures were taken at various intervals.
In 1887, the German botanist Julius von Sachs was the first to examine whether phototropism could be stimulated by particular colors of light (2). By using both colored glass and solutions to illuminate plants with different wavelengths of light, Sachs found that blue light was the most effective.
Phototropins and Blue Light Sensing
Light is one of the most important environmental cues controlling plant development, and is achieved through a suite of photoreceptor proteins. Like photoreceptors associated with our vision, plant photosensors can detect the presence, intensity, direction and color of light, and in turn, utilize this information to direct their growth. To date, four different types of photoreceptors have been identified in plants (3, 4). Among them is a small family of proteins known as the phototropins, which are activated specifically by UV/blue wavelengths of light (5, 6). The photoactivation of these proteins stimulates a range of processes that ultimately optimize the photosynthetic efficiency of plants, including phototropism, after which they were named. For instance, phototropins direct the movement of chloroplasts (Greek for "green maker"), which represent the heart of the photosynthetic machinery as their position within the cell can greatly affect the efficiency of energy production (Figure 2A). Likewise, chloroplasts reside in leaves, which can be viewed simply as solar panels. Leaf positioning and expansion is also directed by the phototropins (Figure 2B, C). Additionally, phototropins control the opening of stomata (Greek for "mouths") pores in the leaf epidermis, which regulate gaseous exchange (Figure 2D). Stomatal opening is important for energy production, as it allows CO2 uptake for photosynthesis. Collectively, these responses serve to enhance the photosynthetic performance of plants and maximize their growth potential (7). Many plant species are able to track the movement of the sun by a process known as heliotropism (Greek for "towards sun"). This photomovement response is also likely mediated by phototropins.
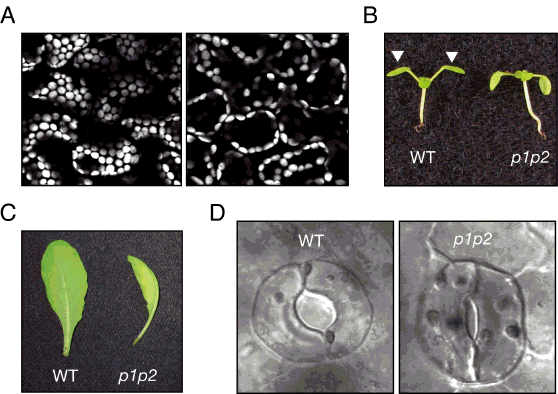
Figure 2. Phototropin-mediated responses in Arabidopsis. (A) Chloroplast positioning in leaf cells of Arabidopsis. Microscope images represent cells viewed from above. Accumulation movement of chloroplasts to the cell surface (left) occurs when leaves are irradiated from above with low intensity blue light. Avoidance movement of chloroplasts (right) to the cell sidewalls occurs when leaves are irradiated with high intensity blue light. (B) Leaf positioning in wild-type (WT) and phototropin-deficient Arabidopsis plants (p1p2), as indicated by the white arrows, in response to irradiation from above. (C) Rosette leaves from mature WT and phototropin-deficient Arabidopsis (p1p2). The leaf from the mutant is unexpanded and curled and therefore lies on its side. (D) Stomatal apertures in light-treated in WT and phototropin-deficient Arabidopsis seedlings (p1p2).
Finding Phototropin
Despite extensive attempts, the molecular identity of the blue light-absorbing photoreceptor responsible for phototropism remained elusive until relatively recently, owing to the availability of genetic methods using the model plant Arabidopsis thaliana (8, 9). Arabidopsis or thale cress, as it is more commonly known, is not the most exciting plant to look at. But its small size and short lifecycle, combined with its plentiful seed production, make it an ideal genetic tool for laboratory work. More importantly, Arabidopsis can be manipulated easily to generate mutants that show altered characteristics. It was the isolation of Arabidopsis mutants altered in phototropism that eventually led to the cloning and characterization of the first phototropin gene (8, 9).
Arabidopsis contains two phototropins referred to as phot1 and phot2 (5, 6). Mutants of Arabidopsis lacking both phototropins lose their phototropic responsiveness (Figure 3). Moreover, genetic and physiological analyses of mutants lacking phot1, phot2 or both photoreceptors demonstrate that they overlap in function to control a number of different photoresponses (5, 6). Both phototropins mediate phototropism, with phot1 acting as the predominant photosensor (10). Indeed, the involvement of phot1 and phot2 in regulating stomatal opening and leaf expansion was not apparent until studies on mutants lacking both proteins were performed (11, 12), again illustrating their functional redundancy.
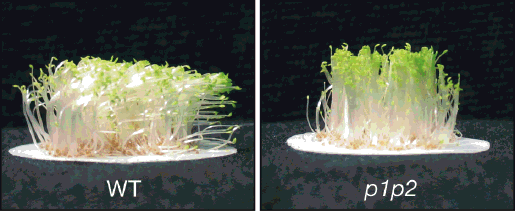
Figure 3. Phototropic response of wild-type Arabidopsis seedlings (WT) and a phototropin-deficient mutant (p1p2) irradiated with blue light from the right.
Similarly, phot1 and phot2 act to induce chloroplast accumulation movement to the upper cell surface under weak light conditions to maximize light capture for photosynthesis (13) (Figure 2A). By contrast, phot2 alone induces chloroplast avoidance movement to the cell sidewalls in bright light to increase mutual shading and prevent photodamage of the photosynthetic machinery under excess light (Figure 2A). Specific functions have also been ascribed to phot1, such as the rapid but transient growth inhibition response of young seedlings upon their emergence from the soil (14).
Phototropin Structure and Light Sensing
The structure of plant phototropins can be separated into two parts: a N-terminal photosensory input region coupled to a C-terminal effector or output region that contains a classic serine/threonine kinase motif (Figure 4A). The N-terminal region comprises two so-called LOV domains, each of which bind the vitamin-B derived cofactor flavin mononucleotide (FMN) as a blue light-absorbing chromophore (15). LOV domains exhibit protein sequence homology to motifs found in a diverse range of eukaryotic and prokaryotic proteins involved in sensing Light, Oxygen, or Voltage, hence the acronym LOV (9). Protein crystallography has shown that the LOV domain consists primarily of five antiparallel (
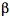 -sheets and two (
-sheets and two (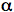 -helices, binding FMN tightly inside an enclosed structure (Figure 4B) (16).
-helices, binding FMN tightly inside an enclosed structure (Figure 4B) (16).
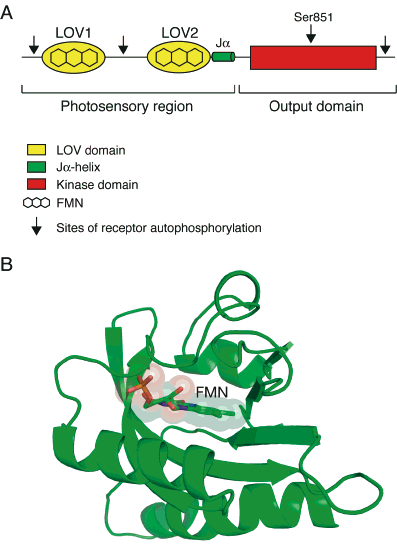
Figure 4. Phototropin structure. (A) Cartoon illustrating the domain structure of phototropin blue light receptors. (B) Structural model of the phototropin LOV2 domain in the dark state. The position of the FMN chromophore is indicated.
LOV domains expressed and purified from Escherichia coli are yellow in color (Figure 5A) owing to their bound flavin cofactor, and are photochemically active in solution as monitored by absorbance spectroscopy (17, 18). In the dark, LOV domains bind FMN non-covalently, forming a spectral species, LOV447, absorbing maximally near 447 nm (Figure 5B). Irradiation of the domain induces the formation of a covalent bond between the C(4a) carbon of the FMN and the sulphur atom of a nearby conserved cysteine residue within the domain (17, 18). Formation of the cysteinyl adduct occurs within microseconds of illumination, and produces a spectral species, LOV390, absorbing maximally near 390 nm. Formation of LOV390 is fully reversible in darkness, and represents the active signaling state that leads to photoreceptor activation. To date, it is still a mystery as to why the phototropins contain two LOV photosensors. Photochemically active LOV2 is necessary for phototropin function (19, 20), while the presence and photochemical reactivity of LOV1 has been shown to be dispensable (19-21). However, LOV1 has been proposed to mediate receptor dimerization and/or modulate the photochemical reactivity of LOV2 (22-24).
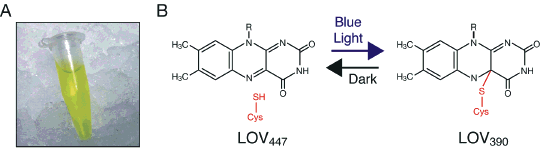
Figure 5. LOV-domain photochemical reactivity. (A) Purified preparation of bacterially expressed LOV2. (B) Schematic representation of LOV-domain photochemistry. Details of the reaction are described in the text.
Phototropin Activation by Light
The current view of phototropin receptor activation is that LOV2 functions as a repressor of the C-terminal kinase domain in the dark, and that this mode of repression is alleviated upon photoexcitation, resulting in receptor autophosphorylation throughout the protein (6, 25) (Figure 6). The photoexcitation of LOV2 leads to the displacement of an (
 -helix from the surface of the domain (26). Unfolding of this (
-helix from the surface of the domain (26). Unfolding of this ( -helix, designated J
-helix, designated J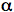 , results in the activation of the C-terminal kinase domain (27). Protein rearrangements within the central (
, results in the activation of the C-terminal kinase domain (27). Protein rearrangements within the central (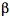 -sheet scaffold have been reported to play a role in propagating the photochemical signal generated within LOV2 domain (28-30) to bring about protein changes at the surface, which are necessary for the activation of the C-terminal kinase domain and autophosphorylation of specific serine/threonine residues (21, 31). Light-activated phototropin can return to its non-phosphorylated state upon incubation in darkness (32). This recovery process involves dephosphorylation of the receptor by an as yet unidentified protein phosphatase.
-sheet scaffold have been reported to play a role in propagating the photochemical signal generated within LOV2 domain (28-30) to bring about protein changes at the surface, which are necessary for the activation of the C-terminal kinase domain and autophosphorylation of specific serine/threonine residues (21, 31). Light-activated phototropin can return to its non-phosphorylated state upon incubation in darkness (32). This recovery process involves dephosphorylation of the receptor by an as yet unidentified protein phosphatase.
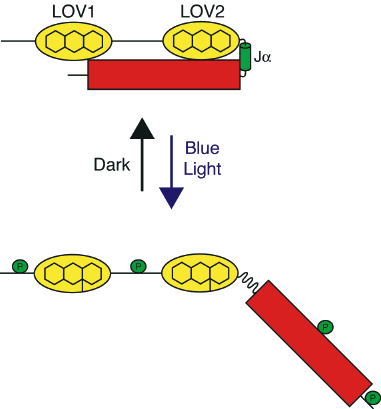
Figure 6. Phototropin activation by blue light. In the dark, phototropin is unphosphorylated and inactive. The absorption of light invokes a photochemical reaction within the LOV domains. Photoexcitation of the main light sensor, LOV2, causes a disordering of the J-helix, and the activation of the C-terminal kinase domain, which consequently leads to autophosphorylation of the photoreceptor. The relative positions of phosphorylation sites are indicated.
Autophosphorylation at a conserved serine residue (Ser851) within the kinase domain of Arabidopsis phot1 is essential for receptor signaling (31) (Figure 4). Although sites of phototropin autophosphorylation have been mapped upstream of LOV2 (21, 31), there is still no information as to the their functional consequences. A truncated version of Arabidopsis phot1, comprising only LOV2 and the C-terminal kinase domain, is functionally active in transgenic Arabidopsis lacking both phot1 and phot2, implying that the phosphorylation of sites upstream of LOV2 is not required for receptor signaling (21).
In darkness, phototropins are typically associated with the plasma membrane, but a small fraction of the receptor pool is rapidly internalized (within minutes) upon blue light irradiation (33, 34). One consequence of autophosphorylation may therefore be to promote receptor dissociation from the plasma membrane, and the desensitizing of the photosensory system analogous to other receptor kinase-based signaling systems. Another consequence of phototropin autophosphorylation is to mediate 14-3-3 binding (35). 14-3-3 proteins are key regulators of protein function in eukaryotes, and preferentially bind to phospho-serine/threonine-containing motifs. However, mutation of the phosphorylation sites required for 14-3-3 binding, located in the peptide region between LOV1 and LOV2, do not appear to perturb the functionality of Arabidopsis phot1 (31). The biological significance of 14-3-3 binding to phototropins therefore awaits further investigation.
Phototropin Signaling
So far, no endogenous substrate for phototropin kinase activity has been identified other than the receptors themselves. Nonetheless, a number of phototropin-interacting proteins have been isolated. Non-Phototropic Hypocotyl 3 (NPH3) is a novel protein that directly interacts with phot1 (36). NPH3 is thought to serve as a protein scaffold to assemble components of a phototropin receptor complex. Arabidopsis mutants lacking NPH3 fail to show phototropism, demonstrating that NPH3 is essential for this response (36). In addition, NPH3 is required for optimal leaf positioning and leaf flattening in Arabidopsis (12). A protein closely related to NPH3, known as Root Phototropism 2 (RPT2) also interacts with phot1, and is required for both phototropism and stomatal opening by blue light (37, 38).
Phototropism (Figure 1) ultimately results from an increase in growth on the shaded side of the stem owing to an accumulation of the plant growth hormone auxin. As light passes through the stem, it becomes progressively diffracted, thereby generating a gradient of phototropin activation across the organ, the highest level of activity occurring on the irradiated side. The formation of this biochemical gradient underlies the directionality of the phototropic response (39). Presently, little information is available regarding how a differential stimulation of phototropin activity across the stem results in an accumulation of auxin on the shaded side of the stem, but likely affects the localization and activity of specific proteins required for auxin transport (40, 41).
Phototropin signaling is undoubtedly complex, given the range of responses that these photoreceptors mediate. Increases in cytosolic calcium concentrations appear to serve as a signaling messenger downstream from receptor activation (42), particularly in the rapid growth-inhibition response that occurs in seedlings emerging from the soil (43). Moreover, cytoskeletal changes are important for both blue light-induced chloroplast movements and stomatal opening (44). A major challenge for future studies will be to unravel these complex signaling networks and to identify the molecular basis of the components involved.
Other LOV-Containing Proteins
In some plants species, including the fern Adiantum capillus-veneris, phototropism and chloroplast movement is induced by red light as well as blue. Adiantum contains a novel dual red/blue light-sensing photoreceptor known as neochrome, comprising of a red light-absorbing phytochrome photosensory domain fused to the N-terminus of an entire phototropin receptor (45). The presence of such a hybrid photoreceptor is proposed to enhance light sensitivity and aid the prevalence of species such as ferns in low light conditions typically found under the canopy of dense forests. Similarly, the photoactivation of red light-absorbing phytochromes is known to enhance blue light-induced phototropism in Arabidopsis (46). However, this phototropic enhancement results from an interaction between separate phytochrome and phototropin receptor systems that likely involves Phytochrome Kinase Substrate (PKS) proteins (47).
Recently, a second LOV-containing photoreceptor family has been characterized in Arabidopsis. Arabidopsis contains three single LOV-containing proteins known as the Zeitlupe (ZTL) family that play important roles in regulating the targeted degradation of components associated with circadian clock function and flowering in a light-dependent manner (48, 49). Furthermore, single LOV-containing proteins are found in other organisms, besides plants. For example, White Collar-1 (WC-1) is a photoreceptor that mediates phototropism in addition to other photoresponses in Neurospora crassa and related fungi (50). Aureochromes are LOV-based transcription factors that mediate photomorphogenesis in photosynthetic algal species living in aquatic environments (51). Surprisingly, LOV-sensing motifs are present in a large number of otherwise very different bacterial proteins that are activated specifically by blue light. Such proteins are found in eubacteria, cyanobacteria and even archea (52). The presence of LOV-containing proteins throughout various kingdoms of life therefore demonstrates that this photosensory module has been conserved throughout evolution. The number and diversity of the LOV-containing proteins found throughout nature is likely to increase as more genome sequences become available. A formidable task will be to elucidate the functions of these bacterial photosensory proteins
Application of the LOV Photosensor
The study of phototropins and related proteins has provided enormous insights into the photochemical and molecular events underlying blue light sensing not only in plants, but also fungi and bacteria. The LOV domain therefore appears to represent an evolutionary conserved input motif that functions to regulate a wide range of output activities including kinases, phosphodiesterases and DNA binding (53). This has prompted research into establishing whether the LOV photosensor can be exploited to generate novel molecular light switches with desirable activities for cell biological or medical applications. Recent work does indeed indicate that the LOV2 photosensory region of plant phototropins can be used effectively as a general regulator of protein activity to produce synthetic light switchable enzymes (54). Such LOV-based molecular switches would offer advantages over ligand-activated systems, because induction requires a non-invasive stimulus (i.e., blue light).
In addition to exploiting its light sensing mechanism, researchers have now begun to take advantage of the fluorescent properties inherent to the LOV domain (55). When irradiated with blue light, LOV domains expressed and purified from E. coli emit a strong green fluorescence, owing to the bound FMN chromophore (Figure 7A). Protein engineering has been employed to improve the fluorescent properties of the LOV domain, such that it can be used successfully as a fluorescent reporter of protein dynamics in living cells (Figure 7B).
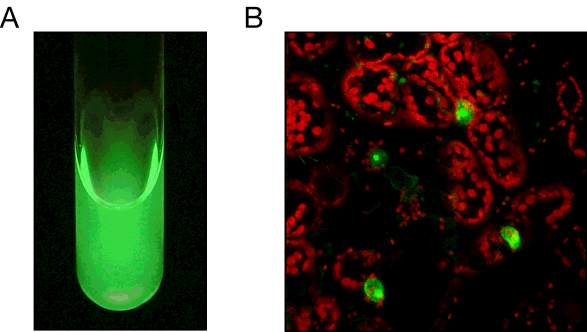
Figure 7. Fluorescent properties of the LOV domain. (A) Solution of purified LOV2 viewed under UV radiation (365 nm). (B) Nuclear fluorescence imaging of Arabidopsis histone 2B in tobacco epidermal cells, as monitored by its fusion to an engineered LOV domain with improved fluorescent properties. LOV fluorescence (green) was visualized by excitation with blue light. Chloroplast autofluorescence is shown in red
While fluorescent proteins based on Green Fluorescent Protein (GFP) from the jellyfish Aequorea victoria are likely to remain the main choice of cell biologists for the immediate future, LOV-based fluorescent probes are likely to provide attractive alternatives for specific applications where the current genetically encoded technologies fall short. For instance, LOV-based fluorescent proteins have been used successfully to monitor bacterial cell populations under anaerobic conditions (55), which is problematic when using GFP. Furthermore, LOV-based proteins can function as superior reporters to GFP for monitoring local and systemic infections of plant RNA viruses, due to their smaller size and reduced genetic load (Figure 8). Thus, the smaller size of LOV-based proteins over GFP and its relatives will likely extend the range of plant and animal viruses that can be fluorescently tagged in vivo, and enhance the study of intercellular protein trafficking.

Figure 8. LOV out performs GFP as a fluorescent reporter of plant virus infection. Upper leaves of tobacco showing the degree of systemic movement of Tobacco mosaic virus (TMV) expressing either an engineered LOV domain or GFP. TMV-LOV shows systemic spread and unloads from all major vein classes, spreading into neighbouring ground tissue (left), whereas TMV-GFP does not cause systemic infection (right).
Conclusions
In the last decade, progress in understanding the photochemical and properties of plant phototropins, in addition to their functional roles has been enormous. Identification of the LOV domain as a blue-light-sensing module, and deciphering its structure and photochemical reactivity represents a major advance. Moreover, the presence of LOV-containing proteins throughout various kingdoms of life clearly demonstrates that this functional blue light sensor is not only restricted to plants, but has been conserved throughout evolution, greatly expanding the potential directions for future research. Work has already begun to exploit the photochemical properties of these small molecular light switches, and will undoubtedly yield further exciting advances in the years to come.
References
1. Darwin, C. (1880) Power of movement in plants (John Murray: London).
2. Sachs, (von) J. (1887) Lectures on the physiology of plants (Clarendon: Oxford).
3. Chen, M., Chory, J. and Fankhauser, C. (2004) Light signal transduction in higher plants. Annu. Rev. Genet. 38, 87-117.
4. Banerjee, R. and Batschauer, A. (2005) Plant blue-light receptors. Planta 220, 498-502.
5. Briggs, W.R. and Christie, J.M. (2002) Phototropin 1 and phototropin 2: Two versatile plant blue-light receptors. Trends Plant Sci. 7, 204-209.
6. Christie, J.M. (2007) Phototropin blue-light receptors, Annu. Rev. Plant Biol., 59, 21-45.
7. Takemiya, A., Inoue, S., Doi, M., Kinoshita, T. and Shimazaki, K. (2005) Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17, 1120-1127.
8. Liscum, E. and Briggs, W.R. (1995) Mutations in the NPH1 locus disrupt th perception of phototropic stimuli. Plant Cell 7, 473-485.
9. Huala, E., Oeller, P.W., Liscum, E., Han, I.-S., Larsen, E. and Briggs, W.R. (1997) Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120-2123.
10. Sakai, T., Kagawa, T., Kasahara, M., Swartz, T.E., Christie, J.M. et al. (2001) Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98, 6969-6974.
11. Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M. and Shimazaki, K. (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656-660.
12. Inoue, S., Kinoshita, T., Takemiya, A., Doi, M. and Shimazaki, K. (2008) Leaf positioning of Arabidopsis in responses to blue light. Mol. Plant 1, 15-26.
13. Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S. et al. (2001) Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138-2141.
14. Folta, K.M. and Spalding, E. (2001) Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26, 471-478.
15. Christie, J.M., Salomon, M., Nozue, K., Wada, M. and Briggs, W.R. (1999) LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96, 8779-8783.
16. Crosson, S. and Moffat, K. (2001) Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98, 2995-3000.
17. Salomon, M. Christie, J.M., Knieb, E., Lempert, U. and Briggs, W.R. (2000) Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochem. 39, 9401-9410.
18. Swartz, T.E., Corchnoy, S.B., Christie, J.M., Lewis, J.W., Szundi, I. et al. (2001) The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276: 36493-36500.
19. Christie, J.M., Swartz, T.E., Bogomolni, R.A. and Briggs, W.R. (2002) Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32, 205-219.
20. Cho, H.-Y., Tseng, T.S., Kaiserli, E., Sullivan., S., Christie, J.M. et al. (2007) Physiological roles of the LOV domains of phototropin 1 and phototropin 2 in Arabidopsis thaliana. Plant Physiol. 143, 517-529.
21. Sullivan, S., Thomson, C.E., Lamont, D.J., Jones, M.A. and Christie, J.M. (2008) In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototopin 1. Mol. Plant 1, 178-194.
22. Salomon, M., Lempert, U. and Rudiger, W. (2004) Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 572, 8-10.
23. Matsuoka, D. and Tokutomi, S. (2005) Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. USA 102, 13337-13342.
24. Nakasako, M., Zikihara, K., Matsuoka, D., Katsura, H. and Tokutomi, S. (2008) Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 381, 718-733.
25. Matsuoka, D., Iwata, T., Zikihara, K., Kandori, H. and Tokutomi, S. (2007) Primary processes during the light-signal transduction of phototropin. Photochem. Photobiol. 83, 121-130.
26. Harper, S.M., Neil, L.C. and Gardner, K.H. (2003) Structural basis of a phototropin light switch. Science 301, 1541-1544.
27. Harper, S.M., Christie, J.M. and Gardner, K.H. (2004) Disruption of the LOV-J alpha helix interaction activates phototropin kinase activity. Biochem. 43, 16184-16192.
28. Nozaki, D. Iwata, T., Ishikawa, T., Todo, T., Tokutomi, S. et al. (2004) Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome 3. Biochem. 43, 8373-8379.
29. Iwata, T., Nozaki, D., Tokutomi, S. and Kandori, H. (2005) Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome 3. Biochem. 44, 7427-7434.
30. Jones, M.A., Feeney, K.A., Kelly, S.M. and Christie, J.M. (2007) Mutational analysis of phototropin 1 provides insights into the mechanisms underlying LOV2 signal transmission. J. Biol. Chem. 282, 6405-6414.
31. Inoue, S., Kinoshita, T., Matsumoto, M., Nakayama, K., Doi, M. et al. (2008) Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 105, 5626-5631.
32. Short, T.W. and Briggs, W.R. (1990). Characterization of a rapid, blue light-Mediated change in detectable phosphorylation of a plasma membrane protein from etiolated pea (Pisum sativum L.) seedlings. Plant Physiol 92, 179-185.
33. Sakamoto, K. and Briggs, W.R. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell, 14, 1723-1735.
34. Kong, S.G., Suzuki, T., Tamura, K., Mochizuki, N., Hara-Nishimura, I. et al. (2006) Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 45, 994-1005.
35. Kinoshita, T., Emi, T., Tominaga, M., Sakamoto, K., Shigenaga, A. et al. (2003). Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiol. 133, 1453-1463.
36. Motchoulski, A. and Liscum, E. (1999) Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961-964.
37. Sakai, T., Wada, T., Ishiguro, S. and Okada, K. (2000) RPT2: A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12, 225-236.
38. Inada, S., Ohgishi, M., Mayama, T., Okada, K. and Sakai, T. (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16, 887-896.
39. Salomon, M., Zacherl, M. and Rudiger, W. (1997) Phototropism and protein phosphorylation in higher plants: unilateral blue bight irradiation generates a directional gradient of protein phosphorylation across the oat coleoptile. Botanica Acta 110, 214-216.
40. Blakeslee, J.J., Bandtopadhyay, A., Peer, W.A., Makam, S.N. and Murphy, A.S. (2004) Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134, 28-31.
41. Noh, B., Bandtopadhyay, A., Peer, W.A., Spalding, E.P. and Murphy, A.S. (2003) Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423, 999-1002.
42. Harada, A., Sakai, T. and Okada, K. (2003) Phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc. Natl. Acad. Sci. USA 100, 8583-8588.
43. Folta, K.M., Lieg, E.J., Durham, T. and Spalding, E.P. (2003) Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol. 133, 1464-1470.
44. Wada, M., Kagawa, T. and Sato, Y. (2003) Chloroplast movement. Annu. Rev. Plant Biol. 54, 455-468.
45. Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H. et al. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95, 15826-15830.
46. Lariguet, P. and Fankhauser, C. (2004) Hypocotyl growth orientation in blue light is determined by phytochrome A inhibition of gravitropism and phototropin promotion of phototropism. Plant J. 40, 826-834.
47. Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U.V., Trevisan M. et al. (2006) PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103, 10134-10139.
48. Kim, W.-Y., Fujiwara, S., Suh, S.-S., Kim, J., Kim, Y. et al. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356-360.
49. Sawa, M., Nusinow, D.A., Kay, S.A. and Imaizumi, T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261-265.
50. Ballario, P., Vittorioso, P., Magrelli, A., Talora, C., Cabibbo, A. et al. (1996) White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15, 1650-1657.
51. Takahashi, F., Yamagata, D., Ishikawa, M., Fukamatsu, Y., Ogura, Y. et al. (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. USA 104, 19625-19630.
52. Briggs, W.R. (2007) The LOV domain: a chromophore module servicing multiple photoreceptors. J. Biomed. Sci. 14, 499-504.
53. Crosson, S., Rajagopal, S. and Moffat, K. (2003) The LOV domain family: Photoresponsive signaling modules coupled to divers output domains. Biochem. 42, 2-10.
54. Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauss, U. et al. (2007) Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443-445.
55. Strickland, D., Moffat, K. and Sosnick, T. (2008) Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. USA 105, 10709-10714.
12/2/08